Nuclear talin-1 provides a bridge between cell adhesion and gene expression
- PMID: 39898029
- PMCID: PMC11787672
- DOI: 10.1016/j.isci.2025.111745
Nuclear talin-1 provides a bridge between cell adhesion and gene expression
Abstract
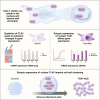
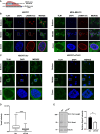
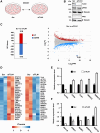
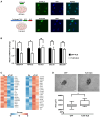
Talin-1 (TLN1) is best known to activate integrin receptors and transmit mechanical stimuli to the actin cytoskeleton at focal adhesions. However, the localization of TLN1 is not restricted to focal adhesions. By utilizing both subcellular fractionations and confocal microscopy analyses, we show that TLN1 localizes to the nucleus in several human cell lines, where it is tightly associated with the chromatin. Importantly, small interfering RNA (siRNA)-mediated depletion of endogenous TLN1 triggers extensive changes in the gene expression profile of human breast epithelial cells. To determine the functional impact of nuclear TLN1, we expressed a TLN1 fusion protein containing a nuclear localization signal. Our findings revealed that the accumulation of nuclear TLN1 alters the expression of a subset of genes and impairs the formation of cell-cell clusters. This study introduces an additional perspective on the canonical view of TLN1 subcellular localization and function.
Keywords: cell biology; cellular physiology; molecular biology; molecular interaction; properties of biomolecules.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

