Molecular assessment of oyster microbiomes and viromes reveals their potential as pathogen and ecological sentinels
- PMID: 39898315
- PMCID: PMC11786891
- DOI: 10.1016/j.onehlt.2025.100973
Molecular assessment of oyster microbiomes and viromes reveals their potential as pathogen and ecological sentinels
Abstract
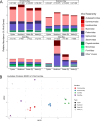
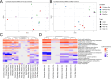
Oyster aquaculture world-wide is a booming industry that can provide many benefits to coastal habitats, including economic, ecosystem-level, and cultural benefits. Oysters present several risks for human consumption, including transmission of parasites, and bacterial and viral pathogens. Oyster microbiomes are well-defined, but their connection to the incidence of pathogens, humans or others, is unclear. Furthermore, viruses associated with oysters are largely unknown, and their connection to humans, animals, and ecosystem health has not been explored. Here, we employed a One Health framework and modern molecular techniques, including 16S rRNA amplicon and metagenomic sequencing, to identify links between changes in the microbial and viral communities associated with oysters and the incidence of pathogens detected in oyster tissues and their surrounding environments. In addition, we adapted the BioFire® FilmArray®, commonly used in hospitals, to determine the presence of human pathogens within the sampled oysters. We detected known human pathogens in 50 % of the oysters tested. Within the genomic datasets, we noted that pathogens of humans, animals, and plants in oysters were shared with the nearby water and sediments, suggesting a sink-source dynamic between the oysters and their surroundings. 16S rRNA gene analysis revealed that while oysters share common microbial constituents with their surrounding environments, they enrich for certain bacteria such as Mycoplasmatales, Fusobacteriales, and Spirochaetales. On the contrary, we found that oyster viromes harbored the same viruses in near equal relative abundances as their surrounding environments. Our results show how oysters could be used not only to determine the risk of human pathogens within coastal estuaries but also how oyster viruses could be used as ecosystem-level sentinels.
Keywords: Aquaculture; Food safety; Microbiome; Oysters; Viruses.
© 2025 The Authors. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Host and Water Microbiota Are Differentially Linked to Potential Human Pathogen Accumulation in Oysters.Appl Environ Microbiol. 2023 Jul 26;89(7):e0031823. doi: 10.1128/aem.00318-23. Epub 2023 Jun 15. Appl Environ Microbiol. 2023. PMID: 37318344 Free PMC article.
-
The core microbiome of cultured Pacific oyster spat is affected by age but not mortality.Microbiol Spectr. 2024 Oct 3;12(10):e0003124. doi: 10.1128/spectrum.00031-24. Epub 2024 Aug 20. Microbiol Spectr. 2024. PMID: 39162495 Free PMC article.
-
Oyster aquaculture enhances sediment microbial diversity- Insights from a multi-omics study.bioRxiv [Preprint]. 2023 Nov 13:2023.11.13.566866. doi: 10.1101/2023.11.13.566866. bioRxiv. 2023. PMID: 38014072 Free PMC article. Preprint.
-
From Farm to Fingers: an Exploration of Probiotics for Oysters, from Production to Human Consumption.Probiotics Antimicrob Proteins. 2020 Jun;12(2):351-364. doi: 10.1007/s12602-019-09629-3. Probiotics Antimicrob Proteins. 2020. PMID: 32056150 Review.
-
Microplastic in oysters: A review of global trends and comparison to southern Australia.Chemosphere. 2022 Nov;307(Pt 4):136065. doi: 10.1016/j.chemosphere.2022.136065. Epub 2022 Aug 19. Chemosphere. 2022. PMID: 35995196 Review.
References
-
- Beck M.W., Brumbaugh R.D., Airoldi L., Carranza A., Coen L.D., Crawford C., Defeo O., Edgar G.J., Hancock B., Kay M.C., Lenihan H.S., Luckenbach M.W., Toropova C.L., Zhang G.F., Guo X.M. Oyster reefs at risk and recommendations for conservation, restoration, and management. Bioscience. 2011;61:107–116. doi: 10.1525/bio.2011.61.2.5. - DOI
-
- Beck M.W., Heck K.L., Able K.W., Childers D.L., Eggleston D.B., Gillanders B.M., Halpern B., Hays C.G., Hoshino K., Minello T.J., Orth R.J., Sheridan P.F., Weinstein M.R. The identification, conservation, and management of estuarine and marine nurseries for fish and invertebrates. Bioscience. 2001;51:633–641. doi: 10.1641/0006-3568(2001)051[0633:TICAMO]2.0.CO;2. - DOI
-
- Grabowski J.H., Brumbaugh R.D., Conrad R.F., Keeler A.G., Opaluch J.J., Peterson C.H., Piehler M.F., Powers S.P., Smyth A.R. Economic valuation of ecosystem services provided by oyster reefs. Bioscience. 2012;62:900–909. doi: 10.1525/bio.2012.62.10.10. - DOI
-
- Newell R.I.E. Ecosystem influences of natural and cultivated populations of suspension-feeding bivalve molluscs: a review. J. Shellfish Res. 2004;23:51–61.
-
- Olivier A.V., Jones L., Le Vay L., Christie M., Wilson J., Malham S.K. A global review of the ecosystem services provided by bivalve aquaculture. Rev. Aquac. 2020;12:3–25. doi: 10.1111/raq.12301. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

