Transcription of hepatitis B surface antigen shifts from cccDNA to integrated HBV DNA during treatment
- PMID: 39898797
- PMCID: PMC11910228
- DOI: 10.1172/JCI184243
Transcription of hepatitis B surface antigen shifts from cccDNA to integrated HBV DNA during treatment
Abstract
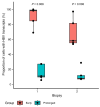
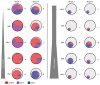
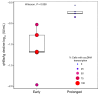
The cornerstone of functional cure for chronic hepatitis B (CHB) is hepatitis B surface antigen (HBsAg) loss from blood. HBsAg is encoded by covalently closed circular DNA (cccDNA) and HBV DNA integrated into the host genome (iDNA). Nucleos(t)ide analogs (NUCs), the mainstay of CHB treatment, rarely lead to HBsAg loss, which we hypothesized was due to continued iDNA transcription despite decreased cccDNA transcription. To test this, we applied a multiplex droplet digital PCR that identifies the dominant source of HBsAg mRNAs to 3,436 single cells from paired liver biopsies obtained from 10 people with CHB and HIV receiving NUCs. With increased NUC duration, cells producing HBsAg mRNAs shifted their transcription from chiefly cccDNA to chiefly iDNA. This shift was due to both a reduction in the number of cccDNA-containing cells and diminished cccDNA-derived transcription per cell; furthermore, it correlated with reduced detection of proteins deriving from cccDNA but not iDNA. Despite this shift in the primary source of HBsAg, rare cells remained with detectable cccDNA-derived transcription, suggesting a source for maintaining the replication cycle. Functional cure must address both iDNA and residual cccDNA transcription. Further research is required to understand the significance of HBsAg when chiefly derived from iDNA.
Keywords: Hepatitis; Infectious disease; Transcription; Virology.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

