Profiling DNA damage in 3D Histology Samples
- PMID: 39899002
- PMCID: PMC7617225
- DOI: 10.1007/978-3-031-16961-8_9
Profiling DNA damage in 3D Histology Samples
Abstract
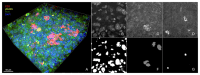
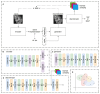
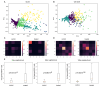
The morphology of individual cells can reveal much about the underlying states and mechanisms in biology. In tumor environments, the interplay among different cell morphologies in local neighborhoods can further improve this characterization. In this paper, we present an approach based on representation learning to capture similarities and subtle differences in cells positive for γH2AX, a common marker for DNA damage. We demonstrate that texture representations using GLCM and VAE-GAN enable profiling of cells in both singular and local neighborhood contexts. Additionally, we investigate a possible quantification of immune and DNA damage response interplay by enumerating CD8+ and γH2AX+ on different scales. Using our profiling approach, regions in treated tissues can be differentiated from control tissue regions, demonstrating its potential in aiding quantitative measurements of DNA damage and repair in tumor contexts.
Keywords: 3D histology; DNA damage; cell morphology; representation learning.
Figures
References
-
- Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467) - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
