Peripheral Transcriptomics in Acute and Long-Term Kidney Dysfunction in SARS-CoV-2 Infection
- PMID: 39899385
- PMCID: PMC12233856
- DOI: 10.34067/KID.0000000727
Peripheral Transcriptomics in Acute and Long-Term Kidney Dysfunction in SARS-CoV-2 Infection
Abstract
Key Points:
AKI in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is linked to mitochondrial dysfunction and cellular stress, highlighting novel biomarkers for severe AKI.
Molecular similarities between AKI in SARS-CoV-2 and sepsis suggest that current therapeutic strategies may be applicable across both conditions.
Molecular changes in acute SARS-CoV-2 infection are tied to long-term inflammation, immune dysregulation, and lasting cardiac and renal dysfunction.
Background: AKI is common in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and coronavirus disease 2019, often leading to long-term kidney dysfunction. However, the transcriptomic features of AKI severity and its long-term effects are underexplored.
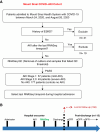
Methods: We performed bulk RNA sequencing on peripheral blood mononuclear cells from hospitalized patients with SARS-CoV-2 infection and complemented these findings with proteomic data from the same cohort. We compared the functional enrichment findings with historical sepsis–AKI data and subsequently examined the association between molecular signatures and long-term kidney function changes.
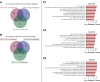
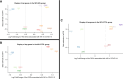
Results: In 283 patients, 57 had mild AKI (stage 1) and 49 had severe AKI (stage 2 or 3). After adjustments for age, sex, severity of infection, and preexisting CKD, we identified 6432 differentially expressed genes in the severe AKI versus control comparison, 840 in the mild AKI versus control comparison, and 1213 in the severe versus mild AKI comparison (false discovery rate <0.05). Common pathways included unfolded protein response, cellular response to stress using eIF2, and IFN-γ–mediated inflammatory response. Severe AKI was linked to pathways involved in mitochondrial dysfunction and endoplasmic reticulum stress. Proteomic analysis confirmed 40 established AKI and inflammation biomarkers, whereas gene-set enrichment of transcription regulators revealed additional biomarkers for severe AKI. Comparison with peripheral blood mononuclear cell transcriptomics from sepsis-related AKI showed significant functional overlap (30%). Analysis of postdischarge eGFR data in 115 patients identified 177 differentially expressed genes for severe AKI versus control, 106 for mild AKI versus control, and 46 for severe versus mild AKI. Key associations included kidney function decline related to carbohydrate and mitochondrial metabolism, inflammatory-response, and cardiovascular regulation.
Conclusions: We demonstrate that severe AKI in SARS-CoV-2 infection is linked to mitochondrial dysfunction and endoplasmic reticulum stress. The functional overlap with sepsis–AKI suggests potential broader therapeutic applicability. Long-term kidney dysfunction is influenced by disruptions in cellular energy metabolism and immune response.
Keywords: AKI; CKD; COVID-19; congestive heart failure; fibrosis; kidney failure; mRNA; mitochondria; oxidative stress; transcriptional profiling.
Conflict of interest statement
Disclosure forms, as provided by each author, are available with the online version of the article at
Figures


Update of
-
Peripheral Transcriptomics in Acute and Long-Term Kidney Dysfunction in SARS-CoV2 Infection.medRxiv [Preprint]. 2023 Oct 27:2023.10.25.23297469. doi: 10.1101/2023.10.25.23297469. medRxiv. 2023. Update in: Kidney360. 2025 Feb 03;6(6):921-936. doi: 10.34067/KID.0000000727. PMID: 37961671 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

