Deletion of sphingosine 1-phosphate receptor 1 in myeloid cells reduces hepatic inflammatory macrophages and attenuates MASH
- PMID: 39899672
- PMCID: PMC12333770
- DOI: 10.1097/HC9.0000000000000613
Deletion of sphingosine 1-phosphate receptor 1 in myeloid cells reduces hepatic inflammatory macrophages and attenuates MASH
Abstract
Background: Immune cell-driven inflammation is a key mediator of metabolic dysfunction-associated steatohepatitis (MASH) progression. We have previously demonstrated that pharmacological sphingosine 1-phosphate (S1P) receptor modulation ameliorates MASH and is associated with attenuated accumulation of intrahepatic macrophage and T-cell subsets. Although S1P receptors are expressed on several immune cell types, given the prominent role of monocyte-derived recruited macrophages in the sterile inflammation of MASH, we hypothesized that deletion of S1P receptor 1 (S1P1) on myeloid cells may ameliorate MASH by reducing the accumulation of proinflammatory monocyte-derived macrophages in the liver.
Methods: The LyzMCre approach was used to generate myeloid cell-specific knockout mice, termed S1pr1MKO. Littermate S1pr1loxp/loxp mice were used as wild-type controls. MASH was established by feeding mice a high-fat, -fructose, and -cholesterol (FFC) diet for 24 weeks, which led to the development of steatohepatitis and MASH-defining cardiometabolic risk factors. Liver injury and inflammation were determined by histological and gene expression analyses. Intrahepatic leukocyte populations were analyzed by mass cytometry and immunohistochemistry.
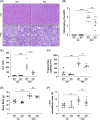
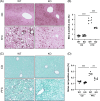
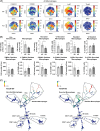
Results: Histological examination demonstrated a reduction in liver inflammatory infiltrates and fibrosis in high-fat, -fructose, and -cholesterol-fed S1pr1MKO compared to wild-type. There was a corresponding reduction in alanine aminotransferase, a sensitive marker for liver injury. As determined by mass cytometry, a significant decrease in recruited macrophages was noted in the livers of high-fat, -fructose, and -cholesterol-fed S1pr1MKO mice compared to wild-type. Gene ontology pathway analysis revealed significant suppression of the peroxisome proliferator-activated receptor gamma and mitogen-activated protein kinase pathways in S1pr1MKO consistent with attenuated MASH in mice.
Conclusions: Deletion of S1P1 in myeloid cells is sufficient to attenuate intrahepatic accumulation of monocyte-derived macrophages and ameliorate murine MASH.
Copyright © 2025 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Association for the Study of Liver Diseases.
Conflict of interest statement
The authors have no conflicts to report.
Figures


References
-
- Nehra V, Angulo P, Buchman AL, Lindor KD. Nutritional and metabolic considerations in the etiology of nonalcoholic steatohepatitis. Dig Dis Sci. 2001;46:2347–2352. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

