RECQ4-MUS81 interaction contributes to telomere maintenance with implications to Rothmund-Thomson syndrome
- PMID: 39900600
- PMCID: PMC11791078
- DOI: 10.1038/s41467-025-56518-1
RECQ4-MUS81 interaction contributes to telomere maintenance with implications to Rothmund-Thomson syndrome
Abstract
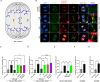
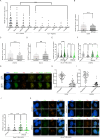
Replication stress, particularly in hard-to-replicate regions such as telomeres and centromeres, leads to the accumulation of replication intermediates that must be processed to ensure proper chromosome segregation. In this study, we identify a critical role for the interaction between RECQ4 and MUS81 in managing such stress. We show that RECQ4 physically interacts with MUS81, targeting it to specific DNA substrates and enhancing its endonuclease activity. Loss of this interaction, results in significant chromosomal segregation defects, including the accumulation of micronuclei, anaphase bridges, and ultrafine bridges (UFBs). Our data further demonstrate that the RECQ4-MUS81 interaction plays an important role in ALT-positive cells, where MUS81 foci primarily colocalise with telomeres, highlighting its role in telomere maintenance. We also observe that a mutation associated with Rothmund-Thomson syndrome, which produces a truncated RECQ4 unable to interact with MUS81, recapitulates these chromosome instability phenotypes. This underscores the importance of RECQ4-MUS81 in safeguarding genome integrity and suggests potential implications for human disease. Our findings demonstrate the RECQ4-MUS81 interaction as a key mechanism in alleviating replication stress at hard-to-replicate regions and highlight its relevance in pathological conditions such as RTS.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Macheret, M. & Halazonetis, T. D. DNA replication stress as a hallmark of cancer. Annu Rev. Pathol.10, 425–448 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

