Nature-inspired platform nanotechnology for RNA delivery to myeloid cells and their bone marrow progenitors
- PMID: 39900620
- PMCID: PMC12014499
- DOI: 10.1038/s41565-024-01847-3
Nature-inspired platform nanotechnology for RNA delivery to myeloid cells and their bone marrow progenitors
Abstract
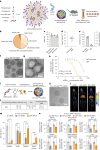
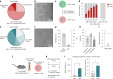
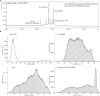
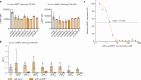
Nucleic acid therapeutics are used for silencing, expressing or editing genes in vivo. However, their systemic stability and targeted delivery to bone marrow resident cells remains a challenge. In this study we present a nanotechnology platform based on natural lipoproteins, designed for delivering small interfering RNA (siRNA), antisense oligonucleotides and messenger RNA to myeloid cells and haematopoietic stem and progenitor cells in the bone marrow. We developed a prototype apolipoprotein nanoparticle (aNP) that stably incorporates siRNA into its core. We then created a comprehensive library of aNP formulations and extensively characterized their physicochemical properties and in vitro performance. From this library, we selected eight representative aNP-siRNA formulations and evaluated their ability to silence lysosomal-associated membrane protein 1 (Lamp1) expression in immune cell subsets in mice after intravenous administration. Using the most effective aNP identified from the screening process, we tested the platform's potential for therapeutic gene silencing in a syngeneic murine tumour model. We also demonstrated the aNP platform's suitability for splice-switching with antisense oligonucleotides and for protein production with messenger RNA by myeloid progenitor cells in the bone marrow. Our data indicate that the aNP platform holds translational potential for delivering various types of nucleic acid therapeutics to myeloid cells and their progenitors.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.G.N. and W.J.M.M. are scientific co-founders of and have equity in Trained Therapeutix Discovery. W.J.M.M. is CSO of Trained Therapeutix Discovery. M.G.N. and W.J.M.M. are scientific co-founders of and have equity in BioTrip. S.R.J.H., T.A., R.Z., H.M.J., P.M.F., W.J.M.M., E.K., and R.v.d.M. are listed as inventors on patent application WO2022268913A1 related to this manuscript. The other authors declare no competing interests.
Figures










References
-
- Kulkarni, J. A. et al. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol.16, 630–643 (2021). - PubMed
-
- Akinc, A. et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol.14, 1084–1087 (2019). - PubMed
-
- Adams, D. et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med.379, 11–21 (2018). - PubMed
MeSH terms
Substances
Grants and funding
- 19681/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organisation for Scientific Research)
- 91818622/ZonMw (Netherlands Organisation for Health Research and Development)
- 101019807/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

