An abundant ginger compound furanodienone alleviates gut inflammation via the xenobiotic nuclear receptor PXR in mice
- PMID: 39900639
- PMCID: PMC11791082
- DOI: 10.1038/s41467-025-56624-0
An abundant ginger compound furanodienone alleviates gut inflammation via the xenobiotic nuclear receptor PXR in mice
Erratum in
-
Author Correction: An abundant ginger compound furanodienone alleviates gut inflammation via the xenobiotic nuclear receptor PXR in mice.Nat Commun. 2025 Mar 3;16(1):2133. doi: 10.1038/s41467-025-57509-y. Nat Commun. 2025. PMID: 40032830 Free PMC article. No abstract available.
Abstract
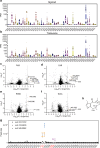
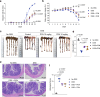
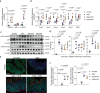
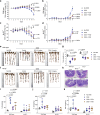
The literature documenting the value of drug-like molecules found in natural products is vast. Although many dietary and herbal remedies have been found to be effective for treating intestinal inflammation, the identification of their active components has lagged behind. In this study, we find that a major ginger component, furanodienone (FDN), is a selective pregnane X receptor (PXR) ligand with agonistic transcriptional outcomes. We show that FDN binds within a sub-pocket of the PXR ligand binding domain (LBD), with subsequent alterations in LBD structure. Using male mice, we show that orally provided FDN has potent PXR-dependant anti-inflammatory outcomes that are colon-specific. Increased affinity and target gene activation in the presence of synergistically acting agonists indicates further opportunities for augmenting FDN activity, efficacy and safety. Collectively, these results support the translational potential of FDN as a therapeutic agent for the treatment and prevention of colonic diseases.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Kuenzig, M. E. et al. Twenty-first century trends in the global epidemiology of pediatric-onset inflammatory bowel disease: systematic review. Gastroenterology162, 1147–1159.e1144 (2022). - PubMed
-
- Burisch, J. et al. The cost of inflammatory bowel disease in high-income settings: a Lancet Gastroenterology & Hepatology Commission. Lancet Gastroenterol. Hepatol.8, 458–492 (2023). - PubMed
MeSH terms
Substances
Grants and funding
- RGPIN480432/Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada)
- PJT-186117/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
- 82204227/National Natural Science Foundation of China (National Science Foundation of China)
- R01 CA222469/CA/NCI NIH HHS/United States
- CRDPJ597036/Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

