Genomic insights into habitat adaptation of Lactobacillus species
- PMID: 39900839
- PMCID: PMC11790720
- DOI: 10.1007/s11274-025-04275-0
Genomic insights into habitat adaptation of Lactobacillus species
Abstract
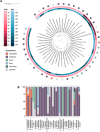
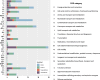
Lactobacillus is one of the most important genera within the lactic acid bacteria group, due to its importance in the food industry and the health field. This diversity can be explained either by their radiation in different environments or by the domestication process in artificial habitats, such as fermented foods. In this study, we performed a comparative genomic analysis of 1020 Lactobacillus genomes, categorizing them into five broad habitats: insects, vertebrates (including humans and animals), vegetables, free-living environments, and dairy products. Utilizing phylogenetic relationships, genomic distances, and gene presence/absence profiles, we identified distinct clustering patterns associated with specific environmental adaptations. Notably, species within the Lactobacillus delbrueckii clade exhibited GC content variations fivefold greater than those observed in other bacterial genera, indicating significant genomic divergence. Insect-associated species showed a strong correlation between genes for carbohydrate utilization and those for amino acid biosynthesis across all habitats. However, individual gene analyses revealed no consistent correlation between habitat adaptation and phylogenetic proximity, suggesting that Lactobacillus employs strain-specific adaptive mechanisms rather than universal genetic markers. Notably, around 50% of the genes associated with specific habitats are hypothetical. Our findings highlight the genomic complexity of Lactobacillus, driven by diverse adaptive strategies, and underscore the need for more comprehensive sampling to fully elucidate the evolutionary dynamics within this important genus.
Keywords: Comparative genomics; Domestication; Habitat Adaptation; Lactobacillus; Pangenomics.
© 2025. The Author(s).
Conflict of interest statement
Decalartaions. Conflict of interest: The authors declare no competing interests.
Figures



References
-
- Albalat R, Cañestro C (2016) Evolution by gene loss. Nat Rev Genet 17(7):379–391. 10.1038/nrg.2016.39 - PubMed
-
- Asnicar F, Thomas AM, Beghini F, Mengoni C, Manara S, Manghi P, Zhu Q, Bolzan M, Cumbo F, May U, Sanders JG, Zolfo M, Kopylova E, Pasolli E, Knight R, Mirarab S, Huttenhower C, Segata N (2020) Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nature Commun 11(1):2500. 10.1038/s41467-020-16366-7 - PMC - PubMed
-
- Boscaro V, Kolisko M, Felletti M, Vannini C, Lynn DH, Keeling PJ (2017) Parallel genome reduction in symbionts descended from closely related free-living bacteria. Nature Ecol Evolut 1(8):1160–1167. 10.1038/s41559-017-0237-0 - PubMed
-
- Broadbent JR, Neeno-Eckwall EC, Stahl B, Tandee K, Cai H, Morovic W, Horvath P, Heidenreich J, Perna NT, Barrangou R, Steele JL (2012) Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genomics 13:533. 10.1186/1471-2164-13-533 - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

