Cardiac repair using regenerating neonatal heart tissue-derived extracellular vesicles
- PMID: 39900896
- PMCID: PMC11790877
- DOI: 10.1038/s41467-025-56384-x
Cardiac repair using regenerating neonatal heart tissue-derived extracellular vesicles
Abstract
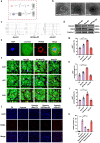
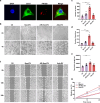
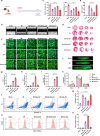
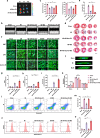
Neonatal mammalian hearts are capable of regenerating by inducing cardiomyocyte proliferation after injury. However, this regenerative capability in adult mammalian hearts almost disappears. Extracellular vesicles (EVs) have been shown to play vital cardioprotective roles in heart repair. Here, we report that EVs from regenerating neonatal heart tissues, after apical resection surgery (AR-Neo-EVs), exhibit stronger pro-proliferative, anti-apoptotic, and pro-angiogenesis activities than EVs from neonatal mouse cardiac tissues (Neo-EVs), effects which are absent in adult mouse heart-derived EVs (Adu-EVs). Proteomic analysis reveals the expression of Wdr75 protein, a regulator of p53, is higher in AR-Neo-EVs than in Neo-EVs. It is shown the regenerative potential of AR-Neo-EVs is abrogated when Wdr75 is knocked down. We further show that delivery of AR-Neo-EVs by sodium alginate hydrogel microspheres is an effective treatment in myocardial infraction. This work shows the potential of using EVs from regenerating tissue to trigger protective and regenerative mechanisms.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


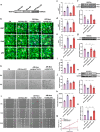
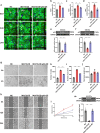

References
-
- Yellon, D. M. & Hausenloy, D. J. Myocardial Reperfusion Injury. N. Engl. J. Med.357, 1121–1135 (2007). - PubMed
-
- Wu, X., Reboll, M. R., Korf-Klingebiel, M. & Wollert, K. C. Angiogenesis after acute myocardial infarction. Cardiovasc. Res.117, 1257–1273 (2021). - PubMed
-
- Heusch, G. Cardioprotection: chances and challenges of its translation to the clinic. Lancet381, 166–175 (2013). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

