Gut microbial dysbiosis associated to diarrheic irritable bowel syndrome can be efficiently simulated in the Mucosal ARtificial COLon (M-ARCOL)
- PMID: 39902883
- PMCID: PMC11796540
- DOI: 10.1080/21655979.2025.2458362
Gut microbial dysbiosis associated to diarrheic irritable bowel syndrome can be efficiently simulated in the Mucosal ARtificial COLon (M-ARCOL)
Abstract
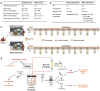
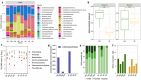
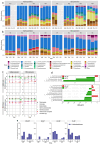
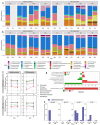
Irritable bowel syndrome (IBS) is a common chronic gastrointestinal disorder, with diarrhea-predominant IBS (IBS-D) as the most frequent subtype. The implication of gut microbiota in the disease's etiology is not fully understood. In vitro gut systems can offer a great alternative to in vivo assays in preclinical studies, but no model reproducing IBS-related dysbiotic microbiota has been developed. Thanks to a large literature review, a new Mucosal ARtifical COLon (M-ARCOL) adapted to IBS-D physicochemical and nutritional conditions was set-up. To validate the model and further exploit its potential in a mechanistic study, in vitro fermentations were performed using bioreactors inoculated with stools from healthy individuals (n = 4) or IBS-D patients (n = 4), when the M-ARCOL was set-up under healthy or IBS-D conditions. Setting IBS-D parameters in M-ARCOL inoculated with IBS-D stools maintained the key microbial features associated to the disease in vivo, validating the new system. In particular, compared to the healthy control, the IBS-D model was characterized by a decreased bacterial diversity, together with a lower abundance of Rikenellaceae and Prevotellaceae, but a higher level of Proteobacteria and Akkermansiaceae. Of interest, applying IBS-D parameters to healthy stools was not sufficient to trigger IBS-D dysbiosis and applying healthy parameters to IBS-D stools was not enough to restore microbial balance. This validated IBS-D colonic model can be used as a robust in vitro platform for studies focusing on gut microbes in the absence of the host, as well as for testing food and microbiota-related interventions aimed at personalized restoration of gut microbiota eubiosis.
Keywords: IBS-D; M-ARCOL; SCFA; bile acids; gut microbiota; in vitro gut model; mucus.
© 2025 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
