ERG responses to high-frequency flickers require FAT3 signaling in mouse retinal bipolar cells
- PMID: 39903280
- PMCID: PMC11793021
- DOI: 10.1085/jgp.202413642
ERG responses to high-frequency flickers require FAT3 signaling in mouse retinal bipolar cells
Abstract
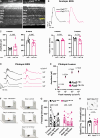
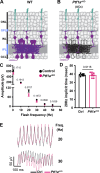
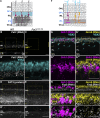
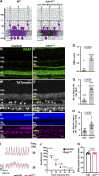
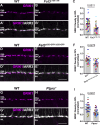
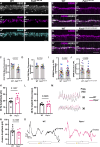
Vision is initiated by the reception of light by photoreceptors and subsequent processing via downstream retinal neurons. Proper circuit organization depends on the multifunctional tissue polarity protein FAT3, which is required for amacrine cell connectivity and retinal lamination. Here, we investigated the retinal function of Fat3 mutant mice and found decreases in both electroretinography and perceptual responses to high-frequency flashes. These defects did not correlate with abnormal amacrine cell wiring, pointing instead to a role in bipolar cell subtypes that also express FAT3. The role of FAT3 in the response to high temporal frequency flashes depends upon its ability to transduce an intracellular signal. Mechanistically, FAT3 binds to the synaptic protein PTPσ intracellularly and is required to localize GRIK1 to OFF-cone bipolar cell synapses with cone photoreceptors. These findings expand the repertoire of FAT3's functions and reveal its importance in bipolar cells for high-frequency light response.
© 2025 Avilés et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures



Update of
-
High temporal frequency light response in mouse retina requires FAT3 signaling in bipolar cells.bioRxiv [Preprint]. 2024 Jun 28:2023.11.02.565326. doi: 10.1101/2023.11.02.565326. bioRxiv. 2024. Update in: J Gen Physiol. 2025 Mar 03;157(2):e202413642. doi: 10.1085/jgp.202413642. PMID: 37961274 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
- International Brain Research Organization
- K99 EY030951/EY/NEI NIH HHS/United States
- Leonard and Isabelle Goldenson Fellowship
- Alice and Joseph Brooks Fund Fellowship
- Lingang Laboratory Startup Fund
- Research to Prevent Blindness
- HHMI/Howard Hughes Medical Institute/United States
- 11241253/Fondecyt de Iniciación Código
- K99EY030951/NH/NIH HHS/United States
- Agencia Nacional de Investigación y Desarrollo
- R01 EY030912/EY/NEI NIH HHS/United States
- The Edward R. and Anne G. Lefler Center
- Harvard Brain Initiative Bipolar Disorders
LinkOut - more resources
Full Text Sources
Research Materials

