Targeting pseudoknots with Cas13b inhibits porcine epidemic diarrhoea virus replication
- PMID: 39903512
- PMCID: PMC11793167
- DOI: 10.1099/jgv.0.002071
Targeting pseudoknots with Cas13b inhibits porcine epidemic diarrhoea virus replication
Abstract
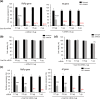
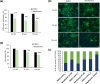
Clustered regularly interspaced short palindromic repeats-associated protein 13 (CRISPR-Cas13), an RNA editing technology, has shown potential in combating RNA viruses by degrading viral RNA within mammalian cells. In this study, we demonstrate the effective inhibition of porcine epidemic diarrhoea virus (PEDV) replication and spread using CRISPR-Cas13. We analysed the sequence similarity of the pseudoknot region between PEDV and severe acute respiratory syndrome coronavirus 2, both belonging to the Coronaviridae family, as well as the similarity of the RNA-dependent RNA polymerase (RdRp) gene region among three different strains of the PED virus. Based on this analysis, we synthesized three CRISPR RNAs (crRNAs) targeting the pseudoknot region and the nonpseudoknot region, each for comparison. In cells treated with crRNA #3 targeting the pseudoknot region, RdRp gene expression decreased by 95%, membrane (M) gene expression by 89% and infectious PEDV titre within the cells reduced by over 95%. Additionally, PED viral nucleocapsid (N) and M protein expression levels decreased by 83 and 98%, respectively. The optimal concentration for high antiviral efficacy without cytotoxicity was determined. Treating cells with 1.5 µg of Cas13b mRNA and 0.5 µg of crRNA resulted in no cytotoxicity while achieving over 95% inhibition of PEDV replication. The Cas13b mRNA therapeutics approach was validated as significantly more effective through a comparative study with merafloxacin, a drug targeting the pseudoknot region of the viral genome. Our results indicate that the pseudoknot region plays a crucial role in the degradation of the PEDV genome through the CRISPR-Cas13 system. Therefore, targeting Cas13b to the pseudoknot offers a promising new approach for treating coronavirus infections.
Keywords: CRISPR RNAs (crRNAs); CRISPR-Cas13b; RNA editing; porcine epidemic diarrhoea virus (PEDV); pseudoknot; viral replication.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


Similar articles
-
Coronavirus Porcine Epidemic Diarrhea Virus Nucleocapsid Protein Interacts with p53 To Induce Cell Cycle Arrest in S-Phase and Promotes Viral Replication.J Virol. 2021 Jul 26;95(16):e0018721. doi: 10.1128/JVI.00187-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34037422 Free PMC article.
-
Field-deployable porcine epidemic diarrhea virus diagnostics utilizing CRISPR-Cas13a.Virulence. 2024 Dec;15(1):2429022. doi: 10.1080/21505594.2024.2429022. Epub 2024 Nov 19. Virulence. 2024. PMID: 39560197 Free PMC article.
-
Pseudoknot-targeting Cas13b combats SARS-CoV-2 infection by suppressing viral replication.Mol Ther. 2023 Jun 7;31(6):1675-1687. doi: 10.1016/j.ymthe.2023.03.018. Epub 2023 Mar 21. Mol Ther. 2023. PMID: 36945774 Free PMC article.
-
In Vitro Antiviral Activities of Salinomycin on Porcine Epidemic Diarrhea Virus.Viruses. 2021 Mar 30;13(4):580. doi: 10.3390/v13040580. Viruses. 2021. PMID: 33808275 Free PMC article.
-
Abrogation of PRRSV infectivity by CRISPR-Cas13b-mediated viral RNA cleavage in mammalian cells.Sci Rep. 2020 Jun 15;10(1):9617. doi: 10.1038/s41598-020-66775-3. Sci Rep. 2020. PMID: 32541822 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

