5-HT orchestrates histone serotonylation and citrullination to drive neutrophil extracellular traps and liver metastasis
- PMID: 39903533
- PMCID: PMC11996869
- DOI: 10.1172/JCI183544
5-HT orchestrates histone serotonylation and citrullination to drive neutrophil extracellular traps and liver metastasis
Abstract
Serotonin (5-HT) is a neurotransmitter that has been linked to tumorigenesis. Whether and how 5-HT modulates cells in the microenvironment to regulate tumor metastasis is largely unknown. Here, we demonstrate that 5-HT was secreted by neuroendocrine prostate cancer (NEPC) cells to communicate with neutrophils and to induce the formation of neutrophil extracellular traps (NETs) in the liver, which in turn facilitated the recruitment of disseminated cancer cells and promoted liver metastasis. 5-HT induced histone serotonylation (H3Q5ser) and orchestrated histone citrullination (H3cit) in neutrophils to trigger chromatin decondensation and facilitate the formation of NETs. Interestingly, we uncovered in this process a reciprocally reinforcing effect between H3Q5ser and H3cit and a crosstalk between the respective writers enzyme transglutaminase 2 (TGM2) and peptidylarginine deiminase 4 (PAD4). Genetic ablation or pharmacological targeting of TGM2, or inhibition of the 5-HT transporter (SERT) with the FDA-approved antidepressant drug fluoxetine reduced H3Q5ser and H3cit modifications, suppressed NET formation, and effectively inhibited NEPC, small-cell lung cancer, and thyroid medullary cancer liver metastasis. Collectively, the 5-HT-triggered production of NETs highlights a targetable neurotransmitter/immune axis that drives liver metastasis of NE cancers.
Keywords: Cell biology; Oncology; Prostate cancer.
Figures

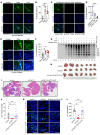
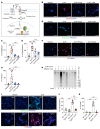
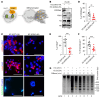




Comment in
- Serotonin sets up neutrophil extracellular traps to promote neuroendocrine prostate cancer metastasis in the liver doi: 10.1172/JCI191687
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

