Identification and validation of TSPAN13 as a novel temozolomide resistance-related gene prognostic biomarker in glioblastoma
- PMID: 39903772
- PMCID: PMC11793784
- DOI: 10.1371/journal.pone.0316552
Identification and validation of TSPAN13 as a novel temozolomide resistance-related gene prognostic biomarker in glioblastoma
Abstract
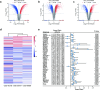
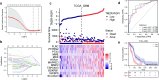
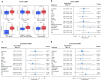
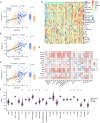
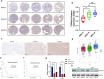
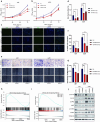
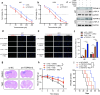
Glioblastoma (GBM) is the most lethal primary tumor of the central nervous system, with its resistance to treatment posing significant challenges. This study aims to develop a comprehensive prognostic model to identify biomarkers associated with temozolomide (TMZ) resistance. We employed a multifaceted approach, combining differential expression and univariate Cox regression analyses to screen for TMZ resistance-related differentially expressed genes (TMZR-RDEGs) in GBM. Using LASSO Cox analysis, we selected 12 TMZR-RDEGs to construct a risk score model, which was evaluated for performance through survival analysis, time-dependent ROC, and stratified analyses. Functional enrichment and mutation analyses were conducted to explore the underlying mechanisms of the risk score and its relationship with immune cell infiltration levels in GBM. The prognostic risk score model, based on the 12 TMZR-RDEGs, demonstrated high efficacy in predicting GBM patient outcomes and emerged as an independent predictive factor. Additionally, we focused on the molecule TSPAN13, whose role in GBM is not well understood. We assessed cell proliferation, migration, and invasion capabilities through in vitro assays (including CCK-8, Edu, wound healing, and transwell assays) and quantitatively analyzed TSPAN13 expression levels in clinical glioma samples using tissue microarray immunohistochemistry. The impact of TSPAN13 on TMZ resistance in GBM cells was validated through in vitro experiments and a mouse orthotopic xenograft model. Notably, TSPAN13 was upregulated in GBM and correlated with poorer patient prognosis. Knockdown of TSPAN13 inhibited GBM cell proliferation, migration, and invasion, and enhanced sensitivity to TMZ treatment. This study provides a valuable prognostic tool for GBM and identifies TSPAN13 as a critical target for therapeutic intervention.
Copyright: © 2025 Wang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Nuclear factor I A promotes temozolomide resistance in glioblastoma via activation of nuclear factor κB pathway.Life Sci. 2019 Nov 1;236:116917. doi: 10.1016/j.lfs.2019.116917. Epub 2019 Oct 12. Life Sci. 2019. PMID: 31614149
-
Aberrant PLAC8 expression characterizes glioblastoma with temozolomide resistance and an immunosuppressive microenvironment.Cancer Lett. 2025 Aug 10;625:217805. doi: 10.1016/j.canlet.2025.217805. Epub 2025 May 19. Cancer Lett. 2025. PMID: 40398706
-
The TNF receptor family member Fn14 is highly expressed in recurrent glioblastoma and in GBM patient-derived xenografts with acquired temozolomide resistance.Neuro Oncol. 2018 Sep 3;20(10):1321-1330. doi: 10.1093/neuonc/noy063. Neuro Oncol. 2018. PMID: 29897522 Free PMC article.
-
Dysregulated lipid metabolism in TMZ-resistant glioblastoma: pathways, proteins, metabolites and therapeutic opportunities.Lipids Health Dis. 2023 Aug 3;22(1):114. doi: 10.1186/s12944-023-01881-5. Lipids Health Dis. 2023. PMID: 37537607 Free PMC article. Review.
-
Post-translational modifications in DNA damage repair: mechanisms underlying temozolomide resistance in glioblastoma.Oncogene. 2025 Jun;44(23):1781-1792. doi: 10.1038/s41388-025-03454-5. Epub 2025 May 26. Oncogene. 2025. PMID: 40419791 Review.
References
-
- Nicholson JG, Fine HA. Diffuse glioma heterogeneity and its therapeutic implications. Cancer Discov. 2021;11(3):575–90. Epub 2021/02/10. doi: 10.1158/2159-8290.CD-20-1474 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

