Reaction Pathway Differentiation Enabled Fingerprinting Signal for Single Nucleotide Variant Detection
- PMID: 39903775
- PMCID: PMC11948007
- DOI: 10.1002/advs.202412680
Reaction Pathway Differentiation Enabled Fingerprinting Signal for Single Nucleotide Variant Detection
Abstract
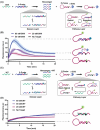
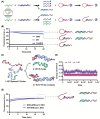
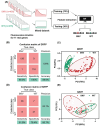
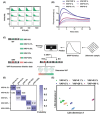
Accurate identification of single-nucleotide variants (SNVs) is paramount for disease diagnosis. Despite the facile design of DNA hybridization probes, their limited specificity poses challenges in clinical applications. Here, a differential reaction pathway probe (DRPP) based on a dynamic DNA reaction network is presented. DRPP leverages differences in reaction intermediate concentrations between SNV and WT groups, directing them into distinct reaction pathways. This generates a strong pulse-like signal for SNV and a weak unidirectional increase signal for wild-type (WT). Through the application of machine learning to fluorescence kinetic data analysis, the classification of SNV and WT signals is automated with an accuracy of 99.6%, significantly exceeding the 80.7% accuracy of conventional methods. Additionally, sensitivity for variant allele frequency (VAF) is enhanced down to 0.1%, representing a ten-fold improvement over conventional approaches. DRPP accurately identified D614G and N501Y SNVs in the S gene of SARS-CoV-2 variants in patient swab samples with accuracy over 99% (n = 82). It determined the VAF of ovarian cancer-related mutations KRAS-G12R, NRAS-G12C, and BRAF-V600E in both tissue and blood samples (n = 77), discriminating cancer patients and healthy individuals with significant difference (p < 0.001). The potential integration of DRPP into clinical diagnostics, along with rapid amplification techniques, holds promise for early disease diagnostics and personalized diagnostics.
Keywords: DNA reaction network; classification; kinetics; machine learning; single‐nucleotide variant detection; variant allele frequency.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Discriminating Interpatient Variabilities of RAS Gene Variants for Precision Detection of Thyroid Cancer.JAMA Netw Open. 2024 May 1;7(5):e2411919. doi: 10.1001/jamanetworkopen.2024.11919. JAMA Netw Open. 2024. PMID: 38758552 Free PMC article.
-
Molecular diagnosis in type I epithelial ovarian cancer.Ginekol Pol. 2017;88(12):692-697. doi: 10.5603/GP.a2017.0123. Ginekol Pol. 2017. PMID: 29303228 Review.
-
Real-Time RT-PCR Allelic Discrimination Assay for Detection of N501Y Mutation in the Spike Protein of SARS-CoV-2 Associated with B.1.1.7 Variant of Concern.Microbiol Spectr. 2022 Feb 23;10(1):e0068121. doi: 10.1128/spectrum.00681-21. Epub 2022 Feb 16. Microbiol Spectr. 2022. PMID: 35170989 Free PMC article.
-
Development and Clinical Validation of Discriminatory Multitarget Digital Droplet PCR Assays for the Detection of Hot Spot KRAS and NRAS Mutations in Cell-Free DNA.J Mol Diagn. 2020 Jul;22(7):943-956. doi: 10.1016/j.jmoldx.2020.04.206. Epub 2020 May 4. J Mol Diagn. 2020. PMID: 32376474
-
Recommendations from the EGAPP Working Group: can testing of tumor tissue for mutations in EGFR pathway downstream effector genes in patients with metastatic colorectal cancer improve health outcomes by guiding decisions regarding anti-EGFR therapy?Genet Med. 2013 Jul;15(7):517-27. doi: 10.1038/gim.2012.184. Epub 2013 Feb 21. Genet Med. 2013. PMID: 23429431
Cited by
-
DNA Molecular Computing with Weighted Signal Amplification for Cancer miRNA Biomarker Diagnostics.Adv Sci (Weinh). 2025 Jun;12(22):e2416490. doi: 10.1002/advs.202416490. Epub 2025 Apr 11. Adv Sci (Weinh). 2025. PMID: 40213938 Free PMC article.
References
-
- a) Abbosh C., Birkbak N. J., Wilson G. A., Jamal‐Hanjani M., Constantin T., Salari R., Le Quesne J., Moore D. A., Veeriah S., Rosenthal R., Nature 2017, 545, 446; - PubMed
- b) Shin H.‐T., Choi Y.‐L., Yun J. W., Kim N. K., Kim S.‐Y., Jeon H. J., Nam J.‐Y., Lee C., Ryu D., Kim S. C., Nat. Commun. 2017, 8, 1377. - PMC - PubMed
-
- a) Kumar S., Thambiraja T. S., Karuppanan K., Subramaniam G., J. Med. Virol. 2022, 94, 1641; - PubMed
- b) Antonelli M., Pujol J. C., Spector T. D., Ourselin S., Steves C. J., Lancet 2022, 399, 2263; - PMC - PubMed
- c) He P., Liu B., Gao X., Yan Q., Pei R., Sun J., Chen Q., Hou R., Li Z., Zhang Y., Nat. Microbiol. 2022, 7, 1635. - PMC - PubMed
-
- a) Tyagi S., Bratu D. P., Kramer F. R., Nat. Biotechnol. 1998, 16, 49; - PubMed
- b) Heyries K. A., Tropini C., VanInsberghe M., Doolin C., Petriv O. I., Singhal A., Leung K., Hughesman C. B., Hansen C. L., Nat. Methods 2011, 8, 649; - PubMed
- c) Schmitt M. W., Kennedy S. R., Salk J. J., Fox E. J., Hiatt J. B., Loeb L. A., Proc. Natl. Acad. Sci. USA 2012, 109, 14508; - PMC - PubMed
- d) Si Y., Wang X., Su X., Weng Z., Hu Q., Li Q., Fan C., Zhang D. Y., Wang Y., Luo S., Angew. Chem., Int. Ed. 2024, 136, e202400551. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
