Lgals3 Promotes Calcium Oxalate Crystal Formation and Kidney Injury Through Histone Lactylation-Mediated FGFR4 Activation
- PMID: 39903812
- PMCID: PMC11947994
- DOI: 10.1002/advs.202413937
Lgals3 Promotes Calcium Oxalate Crystal Formation and Kidney Injury Through Histone Lactylation-Mediated FGFR4 Activation
Abstract
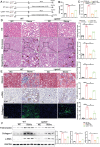
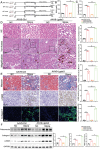
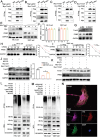
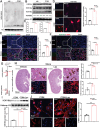
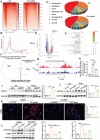
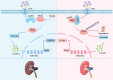
The incidence of kidney stones is increasing worldwide. However, the underlying mechanism of the process of kidney stone formation and the kidney damage caused are not well understood. Here, it is observed that Lgals3, a β-galactoside-binding protein, is significantly increased in tissues with calcium oxalate (CaOx) stones, and in both in vivo and in vitro models. Lgals3 expression is positively correlated with the deposition of CaOx crystals. Knockout of Lgals3 markedly reduces the deposition of CaOx crystal and renal fibrosis in vivo. Furthermore, Lgals3 deficiency decrease the glycolytic rate and lactate production during the process of CaOx deposition and inhibited histone lactylation of H3K18la. Mechanistic studies shows that Lgals3 directly interacted with the key glycolysis protein pyruvate kinase M2 (PKM2) and promoted its expression by modulating E3 ligase Trim21, preventing the ubiquitination of PKM2. Furthermore, H3K18 lactylation promoted CaOx crystal deposition and kidney injury in vivo and in vitro. Lgals3 deficiency inhibites the transcription, activation, and expression of FGFR4 through inhibition of H3K18la. These findings suggest that Lgals3 may play a key role in CaOx stone formation and kidney injury by interacting with PKM2 and promoting both H3K18la-mediated gene transcription and activation.
Keywords: CaOx crystal; Lgals3; epigenetics; histone lactylation; kidney injury.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Singh P., Harris P. C., Sas D. J., Lieske J. C., Nat. Rev. Nephrol. 2022, 18, 224. - PubMed
-
- Thongprayoon C., Krambeck A. E., Rule A. D., Nat. Rev. Nephrol. 2020, 16, 736. - PubMed
-
- Khan S. R., Canales B. K., Dominguez‐Gutierrez P. R., Nat. Rev. Nephrol. 2021, 17, 417. - PubMed
-
- Evan A., Lingeman J., Coe F. L., Worcester E., Kidney Int. 2006, 69, 1313. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
