Unconventional receptor functions and location-biased signaling of the lactate GPCR in the nucleus
- PMID: 39904567
- PMCID: PMC11794946
- DOI: 10.26508/lsa.202503226
Unconventional receptor functions and location-biased signaling of the lactate GPCR in the nucleus
Abstract
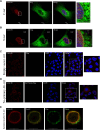
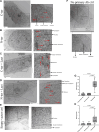
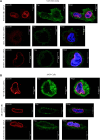
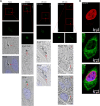
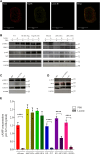
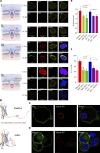
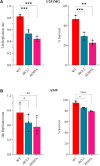
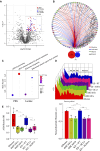
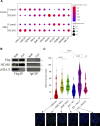
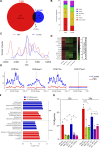
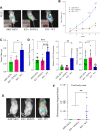
G-protein-coupled receptors (GPCRs) are virtually involved in every physiological process. However, mechanisms for their ability to regulate a vast array of different processes remain elusive. An unconventional functional modality could at least in part account for such diverse involvements but has yet to be explored. We found HCAR1, a multifunctional lactate GPCR, to localize at the nucleus and therein capable of initiating location-biased signaling notably nuclear-ERK and AKT. We discovered that nuclear HCAR1 (N-HCAR1) is directly involved in regulating diverse processes. Specifically, N-HCAR1 binds to protein complexes that are involved in promoting protein translation, ribosomal biogenesis, and DNA-damage repair. N-HCAR1 also interacts with chromatin remodelers to directly regulate gene expression. We show that N-HCAR1 displays a broader transcriptomic signature than its plasma membrane counterpart. Interestingly, exclusion of HCAR1 from the nucleus has the same effect as its complete cellular depletion on tumor growth and metastasis in vivo. These results reveal noncanonical functions for a cell nucleus-localized GPCR that are distinct from traditional receptor modalities and through which HCAR1 can participate in regulating various cellular processes.
© 2025 Mohammad Nezhady et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Boleslavska B, Oravetzova A, Shukla K, Nascakova Z, Ibini ON, Hasanova Z, Andrs M, Kanagaraj R, Dobrovolna J, Janscak P (2022) DDX17 helicase promotes resolution of R-loop-mediated transcription–replication conflicts in human cells. Nucleic Acids Res 50: 12274–12290. 10.1093/nar/gkac1116 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
