Multi-omic spatial effects on high-resolution AI-derived retinal thickness
- PMID: 39904976
- PMCID: PMC11794613
- DOI: 10.1038/s41467-024-55635-7
Multi-omic spatial effects on high-resolution AI-derived retinal thickness
Abstract
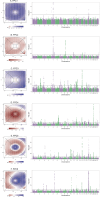
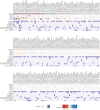
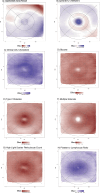
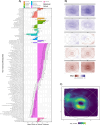
Retinal thickness is a marker of retinal health and more broadly, is seen as a promising biomarker for many systemic diseases. Retinal thickness measurements are procured from optical coherence tomography (OCT) as part of routine clinical eyecare. We processed the UK Biobank OCT images using a convolutional neural network to produce fine-scale retinal thickness measurements across > 29,000 points in the macula, the part of the retina responsible for human central vision. The macula is disproportionately affected by high disease burden retinal disorders such as age-related macular degeneration and diabetic retinopathy, which both involve metabolic dysregulation. Analysis of common genomic variants, metabolomic, blood and immune biomarkers, disease PheCodes and genetic scores across a fine-scale macular thickness grid, reveals multiple novel genetic loci including four on the X chromosome; retinal thinning associated with many systemic disorders including multiple sclerosis; and multiple associations to correlated metabolites that cluster spatially in the retina. We highlight parafoveal thickness to be particularly susceptible to systemic insults. These results demonstrate the gains in discovery power and resolution achievable with AI-leveraged analysis. Results are accessible using a bespoke web interface that gives full control to pursue findings.
© 2025. Crown.
Conflict of interest statement
Competing interests: A.Y.L. reports grants from Santen, personal fees from Genentech, personal fees from US FDA, personal fees from Johnson and Johnson, personal fees from Boehringer Ingelheim, non-financial support from iCareWorld, grants from Topcon, grants from Carl Zeiss Meditec, personal fees from Gyroscope, non-financial support from Optomed, non-financial support from Heidelberg, non-financial support from Microsoft, grants from Regeneron, grants from Amazon, grants from Meta, outside the submitted work; This article does not reflect the views of the US FDA. A.T. reports no direct funding conflicts with the content of the paper. Other disclosures not directly related to the content of the paper: Allergan; Annexon; Apellis; Bayer; 4DMT; Genetech; Heidelberg Engineering; Iveric Bio; Novartis; Oxurion; Roche; VisionAI. C.Y.E. reports receiving financial support from the UK Department of Health through an award made by the National Institute for Health Research to Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology for a Biomedical Research Centre for Ophthalmology. C.Y.E. receives consultant fees from Heidelberg Engineering and Inozyme Pharma unrelated to the scope of the current work. No competing interests reported for M.B., J.O., M.L.G., R.B., B.R.E.A., K.W., V.E.J., L.W.S., S.F., Y.K., Y.W.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

