Fucosylated haptoglobin promotes inflammation via Mincle in sepsis: an observational study
- PMID: 39904983
- PMCID: PMC11794430
- DOI: 10.1038/s41467-025-56524-3
Fucosylated haptoglobin promotes inflammation via Mincle in sepsis: an observational study
Abstract
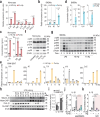
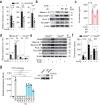
Haptoglobin (Hp) scavenges cell-free hemoglobin and correlates with the prognosis of human sepsis, a life-threatening systemic inflammatory condition. Despite extensive research on Hp glycosylation as a glyco-biomarker for cancers, understanding glycosylated modifications of Hp in sepsis patients (SPs) remains limited. Our study reveals elevated levels of terminal fucosylation at Asn207 and Asn211 of Hp in SP plasma, along with heightened inflammatory responses, compared to healthy controls (trial registration NCT05911711). Fucosylated (Fu)-Hp purified from SPs upregulates inflammatory cytokines and chemokines, along with NLRP3 inflammasome activation. Single-cell RNA sequencing identifies a distinct macrophage-like cell population with increased expressions of inflammatory mediators and FUT4 in response to Fu-Hp. Additionally, Mincle, a C-type lectin receptor, interacts with Fu-Hp to amplify the inflammatory responses and signaling. Moreover, the Hp fucosylation (AAL) level significantly correlates with the levels of inflammatory cytokines in sepsis patients, suggesting that Fu-Hp is clinically relevant. Finally, Fu-Hp treatment significantly enhances the levels of inflammatory cytokines in the plasma and various tissues of mice. Together, our findings reveal a role of Fu-Hp, derived from sepsis patients, in driving inflammation, and suggest that targeting Fu-Hp could serve as a promising intervention for combating sepsis. Trial registration NCT05911711.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- Kristiansen, M. et al. Identification of the haemoglobin scavenger receptor. Nature409, 198–201 (2001). - PubMed
-
- Van Vlierberghe, H., Langlois, M. & Delanghe, J. Haptoglobin polymorphisms and iron homeostasis in health and in disease. Clin. Chim. Acta345, 35–42 (2004). - PubMed
-
- Reinhart, K. et al. Recognizing Sepsis as a Global Health Priority - A WHO Resolution. N. Engl. J. Med377, 414–417 (2017). - PubMed
-
- Harris, E. CDC Introduces Hospital Sepsis Program Guidelines. JAMA330, 1128 (2023). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

