Molecular basis for azetidine-2-carboxylic acid biosynthesis
- PMID: 39905070
- PMCID: PMC11794875
- DOI: 10.1038/s41467-025-56610-6
Molecular basis for azetidine-2-carboxylic acid biosynthesis
Abstract
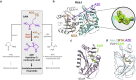
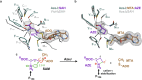
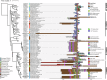
Azetidine-2-carboxylic acid (AZE) is a long-known plant metabolite. Recently, AZE synthases have been identified in bacterial natural product pathways involving non-ribosomal peptide synthetases. AZE synthases catalyse the intramolecular 4-exo-tet cyclisation of S-adenosylmethionine (SAM), yielding a highly strained heterocycle. Here, we combine structural and biochemical analyses with quantum mechanical calculations and mutagenesis studies to reveal catalytic insights into AZE synthases. The cyclisation of SAM is facilitated by an exceptional substrate conformation and supported by desolvation effects as well as cation-π interactions. In addition, we uncover related SAM lyases in diverse bacterial phyla, suggesting a wider prevalence of AZE-containing metabolites than previously expected. To explore the potential of AZE as a proline mimic in combinatorial biosynthesis, we introduce an AZE synthase into the pyrrolizixenamide pathway and thereby engineer analogues of azabicyclenes. Taken together, our findings provide a molecular framework to understand and exploit SAM-dependent cyclisation reactions.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Cornforth, J. W., Redmond, J. W., Eggerer, H., Buckel, W. & Gutschow, C. Asymmetric methyl groups: asymmetric methyl groups, and the mechanism of malate synthase. Nature221, 1212–1213 (1969). - PubMed
-
- Cornforth, J. W., Redmond, J. W., Eggerer, H., Buckel, W. & Gutschow, C. Synthesis and configurational assay of asymmetric methyl groups. Eur. J. Biochem.14, 1–13 (1970). - PubMed
MeSH terms
Substances
Grants and funding
- ANR-20-CE92-0038-01/Agence Nationale de la Recherche (French National Research Agency)
- 325871075-SFB 1309/Deutsche Forschungsgemeinschaft (German Research Foundation)
- KAW 2019.0251/Knut och Alice Wallenbergs Stiftelse (Knut and Alice Wallenberg Foundation)
- NAISS 2023/1-31, NAISS 2023/6-128/Vetenskapsrådet (Swedish Research Council)
- pr83ro/Leibniz-Gemeinschaft (Leibniz Association)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

