Transcriptomic profiles of monocyte-derived macrophages exposed to SARS-CoV-2 VOCs reveal immune-evasion escape driven by delta
- PMID: 39905461
- PMCID: PMC11796281
- DOI: 10.1186/s12967-025-06158-2
Transcriptomic profiles of monocyte-derived macrophages exposed to SARS-CoV-2 VOCs reveal immune-evasion escape driven by delta
Abstract
Background: Since the breakout of COVID-19, the mutated forms of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have shown enhanced rates of transmission and adaptation to humans. The variants of concern (VOC), designated Alpha, Beta, Gamma, Delta, and Omicron emerged independent of one another, and in turn rapidly became dominant. The success of each VOC, as well as the virus fitness, were enabled by altered intrinsic functional properties and, reasonably, to virus antigenicity changes, conferring the ability to evade a primed immune response.
Methods: We analysed the gene expression profiles of monocyte-derived macrophages (MDM) isolated from whole blood of healthy participants exposed to the 5 different SARS-CoV-2 VOC: D614G, Alpha (B.1.1.7), Gamma (P1), Delta (B.1.617.2), and Omicron BA.1 (B.1.1.529), and to the HCoV-OC43 strain, a coronavirus already present in the population before the SARS-CoV-2 pandemic. Whole transcriptome RNA-Seq, for both coding and non-coding RNAs, was then made.
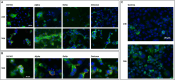
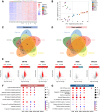
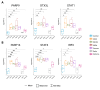
Results: After exposure to the 5 VOC of MDM, we initially assessed the presence of the viral SARS-CoV-2 transcripts to confirm viral entry. We then analysed the RNA-Seq data and observed a significant deregulation of both coding and non-coding RNAs. In particular, our RNA-Seq analysis showed a significant up-regulation of several genes involved in different immunological processes, such as PARP9/PARP14 axes, in macrophages exposed to D614G, Alpha, and Gamma variants. Surprisingly, our data showed that macrophages exposed to the Delta variant exhibited a transcriptional profile more similar to the naïve control group, while macrophages exposed to the Omicron variant showed intermediate differentially expressed genes (DEGs) between the two groups. By checking the canonical markers for M1/M2 differentiation states, we did not observe any expression in macrophages exposed to the Delta variant, suggesting an M0 status, comparable to the naïve control group. Finally, we observed a significant deregulation of 3 main types of non-coding RNAs (ncRNAs): long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and small nucleolar RNAs (snoRNAs), some of which are common to coronaviruses, and some specific to SARS-CoV-2.
Conclusion: The SARS-CoV-2-dependent alteration of the transcriptome of monocyte-derived macrophage (MDM)-infected cells can be linked to the chronological order of the variants' appearance in the human population. Our data suggest an evolution of VOC in modulating the host immune response, with a strong change in pace beginning with the advent of the Delta variant. MDMs exposed to Delta showed a failure in the activation of the adaptive immune response, and this correlates with the more severe symptoms developed by people affected with this SARS-CoV-2 variant.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved (P_175_2021) by the Medical Ethics Committee of IRCCS Policlinico San Matteo of Pavia, and was conducted in accordance with the principles of the Helsinki Declaration. Written informed consent was obtained from all patients prior to enrollment. Consent for publication: Not Applicable. Competing interests: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
Supplementary concepts
Grants and funding
- grant BIAS no. 2020-12371760/Ministero della Salute
- COVID-2020-12371817/Ministero della Salute
- Ricerca Corrente 5x1000 progetto VISION 08069621/Ministero della Salute
- Ricerca Corrente 80728/Ministero della Salute
- Ricerca Corrente/Ministero della Salute
- NextGeneration EU-MUR -National Recovery and Resilience Plan, Mission 4, Component 2 Investment 1.3- Extended Partnership initiative on Emerging Infectious Diseases INF-ACT (project no. PE00000007, CUP B73C22001230006/Ministero dell'Università e della Ricerca
- NextGeneration EU-MUR -PNRR Extended Partnership initiative on Emerging Infectious Diseases INF-ACT (project no. PE00000007, CUP F13C22001220007/Ministero dell'Università e della Ricerca
- Next Generation EU, Missione 4 Componente 1, CUP B53D23016490001/Ministero dell'Università e della Ricerca
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

