A 5-transcript signature for discriminating viral and bacterial etiology in pediatric pneumonia
- PMID: 39906557
- PMCID: PMC11791257
- DOI: 10.1016/j.isci.2025.111747
A 5-transcript signature for discriminating viral and bacterial etiology in pediatric pneumonia
Abstract
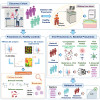
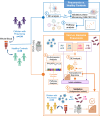
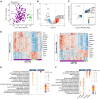
Pneumonia stands as the primary cause of death among children under five, yet current diagnosis methods often result in inadequate or unnecessary treatments. Our research seeks to address this gap by identifying host transcriptomic biomarkers in the blood of children with definitive viral and bacterial pneumonia. We performed RNA sequencing on 192 prospectively collected whole blood samples, including 38 controls and 154 pneumonia cases, uncovering a 5-transcript signature (genes FAM20A, BAG3, TDRD9, MXRA7, and KLF14) that effectively distinguishes bacterial from viral pneumonia (area under the curve (AUC): 0.95 [0.88-1.00]). Initial validation using combined definitive and probable cases yielded an AUC of 0.87 [0.77-0.97], while full validation in a new prospective cohort of 32 patients achieved an AUC of 0.92 [0.83-1.00]. This robust signature holds significant potential to enhance diagnostics accuracy for pediatric pneumonia, reducing diagnostic delays and unnecessary treatments and potentially transforming clinical practice.
Keywords: Body substance sample; Clinical microbiology; Diagnostics; Pediatrics; Transcriptomics.
© 2025 The Author(s).
Conflict of interest statement
The authors have a European patent application related to this work under the identification number EP24382240.
Figures




References
-
- Vos T., Lim S.S., Abbafati C., Abbas K.M., Abbasi M., Abbasifard M., Abbasi-Kangevari M., Abbastabar H., Abd-Allah F., Abdelalim A. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396:1204–1222. - PMC - PubMed
-
- World Health Organization (WHO) 2022. Pneumonia in children.
-
- Haq I.J., Battersby A.C., Eastham K., McKean M. Community acquired pneumonia in children. Br. Med. J. 2017;356:j686. - PubMed
-
- Lynch J.P., III Hospital-acquired pneumonia: risk factors, microbiology, and treatment. Chest. 2001;119:373S–384S. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

