The Japanese encephalitis virus NS1 protein concentrates ER membranes in a cytoskeleton-independent manner to facilitate viral replication
- PMID: 39907281
- PMCID: PMC11915877
- DOI: 10.1128/jvi.02113-24
The Japanese encephalitis virus NS1 protein concentrates ER membranes in a cytoskeleton-independent manner to facilitate viral replication
Abstract
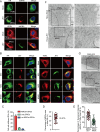
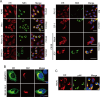
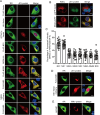
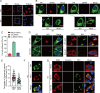
Orthoflaviviruses remodel the endoplasmic reticulum (ER) network to construct replication organelles (ROs) for RNA replication. In this study, we demonstrate that the Japanese encephalitis virus (JEV) NS1 protein concentrates ER membranes in the perinuclear region, which provides a substantial membrane source for viral replication. Subsequently, the virus forms main replication organelles within this membrane-concentrated area to facilitate efficient replication. This process relies on the ER localization signal, glycosylation, dimerization, and membrane-binding sites of the NS1 protein. In conclusion, our study highlights the role of the NS1 protein in the formation of the ROs by JEV, providing new insights into orthoflavivirus replication.IMPORTANCEOrthoflaviviruses use the endoplasmic reticulum (ER) membranes for replication by forming invaginations to assemble the replication organelles. Here, we found that Japanese encephalitis virus (JEV) utilizes the NS1 protein to concentrate a significant number of ER membranes in the perinuclear area, thereby providing a membrane source for viral replication and facilitating the formation of main replication organelles (MROs). This process depends on the ER localization signals of NS1, as well as its glycosylation, dimerization, and membrane-binding sites, but not on the cytoskeleton. In summary, our study highlights how NS1 remodels ER membranes to facilitate the formation of MROs for JEV, thereby accelerating viral replication.
Keywords: ER membranes; Japanese encephalitis virus; NS1; concentration; main replication organelles; viral replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Japanese encephalitis virus NS1 and NS1' proteins induce vimentin rearrangement via the CDK1-PLK1 axis to promote viral replication.J Virol. 2024 May 14;98(5):e0019524. doi: 10.1128/jvi.00195-24. Epub 2024 Apr 24. J Virol. 2024. PMID: 38656209 Free PMC article.
-
The C Terminus of the Core β-Ladder Domain in Japanese Encephalitis Virus Nonstructural Protein 1 Is Flexible for Accommodation of Heterologous Epitope Fusion.J Virol. 2015 Nov 11;90(3):1178-89. doi: 10.1128/JVI.02057-15. Print 2016 Feb 1. J Virol. 2015. PMID: 26559836 Free PMC article.
-
Japanese Encephalitis Virus Induces Apoptosis and Encephalitis by Activating the PERK Pathway.J Virol. 2019 Aug 13;93(17):e00887-19. doi: 10.1128/JVI.00887-19. Print 2019 Sep 1. J Virol. 2019. PMID: 31189710 Free PMC article.
-
Molecular pathogenesis of Japanese encephalitis and possible therapeutic strategies.Arch Virol. 2022 Sep;167(9):1739-1762. doi: 10.1007/s00705-022-05481-z. Epub 2022 Jun 2. Arch Virol. 2022. PMID: 35654913 Free PMC article. Review.
-
Poxvirus membrane biogenesis.Virology. 2015 May;479-480:619-26. doi: 10.1016/j.virol.2015.02.003. Epub 2015 Feb 26. Virology. 2015. PMID: 25728299 Free PMC article. Review.
Cited by
-
TRIM14 restricts tembusu virus infection through degrading viral NS1 protein and activating type I interferon signaling.PLoS Pathog. 2025 May 28;21(5):e1013200. doi: 10.1371/journal.ppat.1013200. eCollection 2025 May. PLoS Pathog. 2025. PMID: 40435148 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

