De novo acquisition of antibiotic resistance in six species of bacteria
- PMID: 39907470
- PMCID: PMC11878088
- DOI: 10.1128/spectrum.01785-24
De novo acquisition of antibiotic resistance in six species of bacteria
Abstract
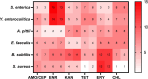
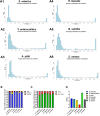
Bacteria can become resistant to antibiotics in two ways: by acquiring resistance genes through horizontal gene transfer and by de novo development of resistance upon exposure to non-lethal concentrations. The importance of the second process, de novo build-up, has not been investigated systematically over a range of species and may be underestimated as a result. To investigate the DNA mutation patterns accompanying the de novo antibiotic resistance acquisition process, six bacterial species encountered in the food chain were exposed to step-wise increasing sublethal concentrations of six antibiotics to develop high levels of resistance. Phenotypic and mutational landscapes were constructed based on whole-genome sequencing at two time points of the evolutionary trajectory. In this study, we found that (1) all of the six strains can develop high levels of resistance against most antibiotics; (2) increased resistance is accompanied by different mutations for each bacterium-antibiotic combination; (3) the number of mutations varies widely, with Y. enterocolitica having by far the most; (4) in the case of fluoroquinolone resistance, a mutational pattern of gyrA combined with parC is conserved in five of six species; and (5) mutations in genes coding for efflux pumps are widely encountered in gram-negative species. The overall conclusion is that very similar phenotypic outcomes are instigated by very different genetic changes. The outcome of this study may assist policymakers when formulating practical strategies to prevent development of antimicrobial resistance in human and veterinary health care.IMPORTANCEMost studies on de novo development of antimicrobial resistance have been performed on Escherichia coli. To examine whether the conclusions of this research can be applied to more bacterial species, six species of veterinary importance were made resistant to six antibiotics, each of a different class. The rapid build-up of resistance observed in all six species upon exposure to non-lethal concentrations of antimicrobials indicates a similar ability to adjust to the presence of antibiotics. The large differences in the number of DNA mutations accompanying de novo resistance suggest that the mechanisms and pathways involved may differ. Hence, very similar phenotypes can be the result of various genotypes. The implications of the outcome are to be considered by policymakers in the area of veterinary and human healthcare.
Keywords: antimicrobial resistance; de novo resistance; resistance genes; resistance mutations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
The Landscape of Phenotypic and Transcriptional Responses to Ciprofloxacin in Acinetobacter baumannii: Acquired Resistance Alleles Modulate Drug-Induced SOS Response and Prophage Replication.mBio. 2019 Jun 11;10(3):e01127-19. doi: 10.1128/mBio.01127-19. mBio. 2019. PMID: 31186328 Free PMC article.
-
QRDR mutations, efflux system & antimicrobial resistance genes in enterotoxigenic Escherichia coli isolated from an outbreak of diarrhoea in Ahmedabad, India.Indian J Med Res. 2011 Aug;134(2):214-23. Indian J Med Res. 2011. PMID: 21911975 Free PMC article.
-
Genome rearrangements in Escherichia coli during de novo acquisition of resistance to a single antibiotic or two antibiotics successively.BMC Genomics. 2018 Dec 27;19(1):973. doi: 10.1186/s12864-018-5353-y. BMC Genomics. 2018. PMID: 30591014 Free PMC article.
-
Mechanisms of drug resistance: quinolone resistance.Ann N Y Acad Sci. 2015 Sep;1354(1):12-31. doi: 10.1111/nyas.12830. Epub 2015 Jul 17. Ann N Y Acad Sci. 2015. PMID: 26190223 Free PMC article. Review.
-
Genetics of antimicrobial resistance.Anim Biotechnol. 2006;17(2):111-24. doi: 10.1080/10495390600957092. Anim Biotechnol. 2006. PMID: 17127523 Review.
Cited by
-
Collateral sensitivity and cross-resistance in six species of bacteria exposed to six classes of antibiotics.Microbiol Spectr. 2025 Aug 5;13(8):e0098325. doi: 10.1128/spectrum.00983-25. Epub 2025 Jun 20. Microbiol Spectr. 2025. PMID: 40539805 Free PMC article.
-
Antimicrobial peptide biological activity, delivery systems and clinical translation status and challenges.J Transl Med. 2025 Mar 7;23(1):292. doi: 10.1186/s12967-025-06321-9. J Transl Med. 2025. PMID: 40055730 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

