Toward Machine Learning Electrospray Ionization Sensitivity Prediction for Semiquantitative Lipidomics in Stem Cells
- PMID: 39907635
- PMCID: PMC11863365
- DOI: 10.1021/acs.jcim.4c02040
Toward Machine Learning Electrospray Ionization Sensitivity Prediction for Semiquantitative Lipidomics in Stem Cells
Abstract
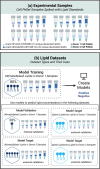
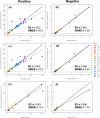
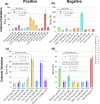
Specificity, sensitivity, and high metabolite coverage make mass spectrometry (MS) one of the most valuable tools in metabolomics and lipidomics. However, translation of metabolomics MS methods to multiyear studies conducted across multiple batches is limited by variability in electrospray ionization response, making batch-to-batch comparisons challenging. This limitation creates an artificial divide between nontargeted discovery work that is broad in scope but limited in terms of absolute quantitation ability and targeted work that is highly accurate but limited in scope due to the need for matched isotopically labeled standards. These issues are often observed in stem cell studies using metabolomic and lipidomic MS approaches, where patient recruitment can be a years-long process and samples become available in discrete batches every few months. To bridge this gap, we developed a machine learning model that predicts electrospray ionization sensitivity for lipid classes that have shown correlation with stem cell potency. Molecular descriptors derived from these lipids' chemical structures are used as model input to predict electrospray response, enabling quantitation by MS with moderate accuracy (semiquantitation). Model performance was evaluated via internal and external validation using cultured cells from various stem cell donors, achieving global percent errors of 40% and 20% for positive and negative electrospray ion modes, respectively. Although this accuracy is typically insufficient for traditional targeted lipidomics experiments, it is sufficient for semiquantitative estimation of lipid marker concentrations across batches without the need for specific chemical standards that many times are unavailable. Furthermore, the precision for model-predicted concentrations was 16.9% for the positive mode and 7.5% for the negative mode, indicating promise for data harmonization across batches. The set of molecular descriptors used by the models described here was able to yield higher accuracy than those previously published in the literature, showing high promise toward semiquantitative lipidomics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Lipid Coverage in Nanospray Desorption Electrospray Ionization Mass Spectrometry Imaging of Mouse Lung Tissues.Anal Chem. 2019 Sep 17;91(18):11629-11635. doi: 10.1021/acs.analchem.9b02045. Epub 2019 Aug 27. Anal Chem. 2019. PMID: 31412198 Free PMC article.
-
Ambient Lipidomic Analysis of Single Mammalian Oocytes and Preimplantation Embryos Using Desorption Electrospray Ionization (DESI) Mass Spectrometry.Methods Mol Biol. 2020;2064:159-179. doi: 10.1007/978-1-4939-9831-9_13. Methods Mol Biol. 2020. PMID: 31565774
-
Microflow Liquid Chromatography Coupled to Multinozzle Electrospray Ionization for Improved Lipidomics Coverage of 3D Clear Cell Renal Cell Carcinoma.Anal Chem. 2025 Mar 11;97(9):5109-5117. doi: 10.1021/acs.analchem.4c06337. Epub 2025 Feb 25. Anal Chem. 2025. PMID: 39998250 Free PMC article.
-
Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-What, how and why?Mass Spectrom Rev. 2017 Nov;36(6):693-714. doi: 10.1002/mas.21492. Epub 2016 Jan 15. Mass Spectrom Rev. 2017. PMID: 26773411 Free PMC article. Review.
-
Enhancing detection and characterization of lipids using charge manipulation in electrospray ionization-tandem mass spectrometry.Chem Phys Lipids. 2020 Oct;232:104970. doi: 10.1016/j.chemphyslip.2020.104970. Epub 2020 Sep 3. Chem Phys Lipids. 2020. PMID: 32890498 Free PMC article. Review.
Cited by
-
Simultaneous Detection of Polar and Nonpolar Molecules by Nano-ESI MS with Plasma Ignited by an Ozone Generator Power Supply.Molecules. 2025 Jun 11;30(12):2546. doi: 10.3390/molecules30122546. Molecules. 2025. PMID: 40572511 Free PMC article.
References
-
- Brachtl G.; Poupardin R.; Hochmann S.; Raninger A.; Jürchott K.; Streitz M.; Schlickeiser S.; Oeller M.; Wolf M.; Schallmoser K.; Volk H.-D.; Geissler S.; Strunk D. Batch Effects during Human Bone Marrow Stromal Cell Propagation Prevail Donor Variation and Culture Duration: Impact on Genotype, Phenotype and Function. Cells 2022, 11 (6), 946.10.3390/cells11060946. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

