Unveiling the native architecture of adult cardiac tissue using the 3D-NaissI method
- PMID: 39907789
- PMCID: PMC11799504
- DOI: 10.1007/s00018-025-05595-y
Unveiling the native architecture of adult cardiac tissue using the 3D-NaissI method
Abstract
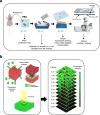
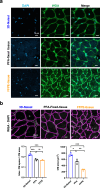
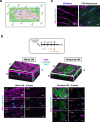
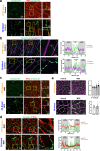
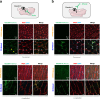
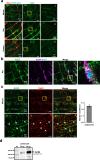
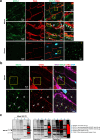
Accurately imaging adult cardiac tissue in its native state is essential for regenerative medicine and understanding heart disease. Current fluorescence methods encounter challenges with tissue fixation. Here, we introduce the 3D-NaissI (3D-Native Tissue Imaging) method, which enables rapid, cost-effective imaging of fresh cardiac tissue samples in their closest native state, and has been extended to other tissues. We validated the efficacy of 3D-NaissI in preserving cardiac tissue integrity using small biopsies under hypothermic conditions in phosphate-buffered saline, offering unparalleled resolution in confocal microscopy for imaging fluorescent small molecules and antibodies. Compared to conventional histology, 3D-NaissI preserves cardiac tissue architecture and native protein epitopes, facilitating the use of a wide range of commercial antibodies without unmasking strategies. We successfully identified specific cardiac protein expression patterns in cardiomyocytes (CMs) from rodents and humans, including for the first time ACE2 localization in the lateral membrane/T-Tubules and SGTL2 in the sarcoplasmic reticulum. These findings shed light on COVID-19-related cardiac complications and suggest novel explanations for therapeutic benefits of iSGLT2 in HFpEF patients. Additionally, we challenge the notion of "connexin-43 lateralization" in heart pathology, suggesting it may be an artifact of cardiac fixation, as 3D-NaissI clearly revealed native connexin-43 expression at the lateral membrane of healthy CMs. We also discovered previously undocumented periodic ring-like 3D structures formed by pericytes that cover the lateral surfaces of CMs. These structures, positive for laminin-2, delineate a specific spatial architecture of laminin-2 receptors on the CM surface, underscoring the pivotal role of pericytes in CM function. Lastly, 3D-NaissI facilitates the mapping of native human protein expression in fresh cardiac autopsies, offering insights into both pathological and non-pathological contexts. Therefore, 3D-NaissI provides unparalleled insights into native cardiac tissue biology and holds the promise of advancing our understanding of physiology and pathophysiology, surpassing standard histology in both resolution and accuracy.
Keywords: ACE2; Cardiac tissue; Cardiomyocytes; Claudin-5; Connexin-43; Fluorescence confocal imaging; Pericytes; SGTL2.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: None declared. Ethical approval: Procedures involving human cardiac samples were conducted at the Department of Forensic Medicine, Centre Hospitalier Universitaire de Toulouse (University of Toulouse, France), adhering strictly to the principles outlined in the Declaration of Helsinki. Furthermore, these procedures received approval in accordance with French legislation from the Agence de Biomédecine under registration number PFS21-015, with explicit consent obtained from the relatives, signifying their non-objection to the sampling process. All animal experiments were performed in accordance with the European directive for the protection of animals used for scientific purpose and were approved of the French CEEA-122 ethical committee (CEEA 122 2015-28). Consent for publication: All the authors have approved and agreed to publish this manuscript.
Figures


References
-
- Shiba Y, Gomibuchi T, Seto T, Wada Y, Ichimura H, Tanaka Y et al (2016) Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 538:388–391 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

