Deterministic genetic barcoding for multiplexed behavioral and single-cell transcriptomic studies
- PMID: 39908076
- PMCID: PMC11798575
- DOI: 10.7554/eLife.88334
Deterministic genetic barcoding for multiplexed behavioral and single-cell transcriptomic studies
Abstract
Advances in single-cell sequencing technologies have provided novel insights into the dynamics of gene expression and cellular heterogeneity within tissues and have enabled the construction of transcriptomic cell atlases. However, linking anatomical information to transcriptomic data and positively identifying the cell types that correspond to gene expression clusters in single-cell sequencing data sets remains a challenge. We describe a straightforward genetic barcoding approach that takes advantage of the powerful genetic tools in Drosophila to allow in vivo tagging of defined cell populations. This method, called
Keywords: D. melanogaster; behavior; genetic barcoding; genetics; genomics; next-generation sequencing; single-cell transcriptomics.
Plain language summary
From delivery to shipping or shopping, barcodes are a part of everyday life. In biological research as well, ‘barcoding’ cells and organisms using specific DNA sequences has been a transformative approach. Such tags can be introduced into the genetic material of cells, allowing scientists to label cell populations or individuals of interest. Here, Mendana et al. investigated how DNA barcoding could be used to cut down the time and cost required to pinpoint a certain population of cells, or of organisms, within a larger group. At present, such efforts often remain labor intensive and costly. For instance, it is now possible for researchers to capture all the genes that are switched on at any given time in individual cells in an organism; however, it is still difficult to then identify which tissue or population of interest a particular cell belongs to. In response, Mendana et al. established a new approach in fruit flies, called TaG-EM, which makes it possible to bypass these limitations by introducing a carefully designed genetic barcode, easily read by DNA sequencers, into the genome of the fly. Further experiments also demonstrated that TaG-EM was valuable at the scale of an organism, to be used in behavioral experiments. Typically, researchers examine how various strains of animals respond to different conditions by testing each group separately; Mendana et al. were able to show that ‘barcoding’ the flies using TaG-EM made it possible to pool these behavioral measurements, as the different groups could then be later quickly identified using their genetic tags. Overall, this new approach should allow researchers using fruit flies to investigate questions around gene expression and behavior in a faster and cheaper way, improving our understanding of a range of biological processes.
© 2023, Blanco Mendana et al.
Conflict of interest statement
JB, MD, LO, BA, JG, DG No competing interests declared
Figures
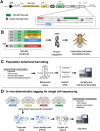

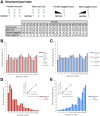
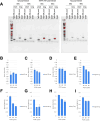

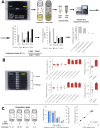





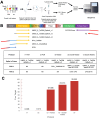
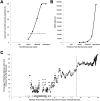
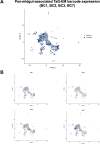
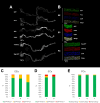

Update of
- doi: 10.1101/2023.03.29.534817
- doi: 10.7554/eLife.88334.1
- doi: 10.7554/eLife.88334.2
References
-
- Alegria AD, Joshi AS, Mendana JB, Khosla K, Smith KT, Auch B, Donovan M, Bischof J, Gohl DM, Kodandaramaiah SB. High-throughput genetic manipulation of multicellular organisms using a machine-vision guided embryonic microinjection robot. Genetics. 2024;226:iyae025. doi: 10.1093/genetics/iyae025. - DOI - PMC - PubMed
-
- Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, Butte AJ, Bhattacharya M. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nature Immunology. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. - DOI - PMC - PubMed
-
- Ariyapala IS, Holsopple JM, Popodi EM, Hartwick DG, Kahsai L, Cook KR, Sokol NS. Identification of split-GAL4 drivers and enhancers that allow regional cell type manipulations of the Drosophila melanogaster intestine. Genetics. 2020;216:891–903. doi: 10.1534/genetics.120.303625. - DOI - PMC - PubMed
-
- Asnicar F, Leeming ER, Dimidi E, Mazidi M, Franks PW, Al Khatib H, Valdes AM, Davies R, Bakker E, Francis L, Chan A, Gibson R, Hadjigeorgiou G, Wolf J, Spector TD, Segata N, Berry SE. Blue poo: impact of gut transit time on the gut microbiome using a novel marker. Gut. 2021;70:1665–1674. doi: 10.1136/gutjnl-2020-323877. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

