Whole-genome sequencing of Acinetobacter baumannii clinical isolates from a tertiary hospital in Terengganu, Malaysia (2011-2020), revealed the predominance of the Global Clone 2 lineage
- PMID: 39908088
- PMCID: PMC11798184
- DOI: 10.1099/mgen.0.001345
Whole-genome sequencing of Acinetobacter baumannii clinical isolates from a tertiary hospital in Terengganu, Malaysia (2011-2020), revealed the predominance of the Global Clone 2 lineage
Abstract
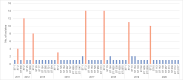
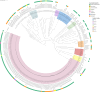
Carbapenem-resistant Acinetobacter baumannii is recognized by the World Health Organization (WHO) as one of the top priority pathogens. Despite its public health importance, genomic data of clinical isolates from Malaysia remain scarce. In this study, whole-genome sequencing was performed on 126 A. baumannii isolates collected from the main tertiary hospital in the state of Terengganu, Malaysia, over a 10-year period (2011-2020). Antimicrobial susceptibilities determined for 20 antibiotics belonging to 8 classes showed that 77.0% (n=97/126) of the isolates were categorized as multidrug resistant (MDR), with all MDR isolates being carbapenem resistant. Multilocus sequence typing analysis categorized the Terengganu A. baumannii clinical isolates into 34 Pasteur and 44 Oxford sequence types (STs), with ST2Pasteur of the Global Clone 2 lineage identified as the dominant ST (n=76/126; 60.3%). The ST2Pasteur isolates could be subdivided into six Oxford STs with the majority being ST195Oxford (n=35) and ST208Oxford (n=17). Various antimicrobial resistance genes were identified with the bla OXA-23-encoded carbapenemase being the predominant acquired carbapenemase gene (n=90/126; 71.4%). Plasmid-encoded rep genes were identified in nearly all (n=122/126; 96.8%) of the isolates with the majority being Rep_3 family (n=121). Various virulence factors were identified, highlighting the pathogenic nature of this bacterium. Only 14/126 (11.1%) of the isolates were positive for the carriage of CRISPR-Cas arrays with none of the prevalent ST2Pasteur isolates harbouring them. This study provided a genomic snapshot of the A. baumannii isolates obtained from a single tertiary healthcare centre in Malaysia over a 10-year period and showed the predominance of a single closely related ST2Pasteur lineage, indicating the entrenchment of this clone in the hospital.
Keywords: Acinetobacter baumannii; GC2 lineage; Malaysia; genome sequencing; multidrug resistant.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




References
-
- Shamsizadeh Z, Nikaeen M, Nasr Esfahani B, Mirhoseini SH, Hatamzadeh M, et al. Detection of antibiotic resistant Acinetobacter baumannii in various hospital environments: potential sources for transmission of Acinetobacter infections. Environ Health Prev Med. 2017;22:44. doi: 10.1186/s12199-017-0653-4. - DOI - PMC - PubMed
-
- Teerawattanapong N, Panich P, Kulpokin D, Na Ranong S, Kongpakwattana K, et al. A systematic review of the burden of multidrug-resistant healthcare-associated infections among intensive care unit patients in southeast Asia: the rise of multidrug-resistant Acinetobacter baumannii. Infect Control Hosp Epidemiol. 2018;39:525–533. doi: 10.1017/ice.2018.58. - DOI - PubMed
-
- Mohd Sazlly Lim S, Zainal Abidin A, Liew SM, Roberts JA, Sime FB. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: a systematic review and meta-analysis. J Infect. 2019;79:593–600. doi: 10.1016/j.jinf.2019.09.012. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

