LitSumm: large language models for literature summarization of noncoding RNAs
- PMID: 39908113
- PMCID: PMC11833236
- DOI: 10.1093/database/baaf006
LitSumm: large language models for literature summarization of noncoding RNAs
Abstract
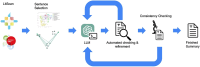
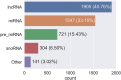
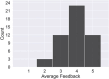
Curation of literature in life sciences is a growing challenge. The continued increase in the rate of publication, coupled with the relatively fixed number of curators worldwide, presents a major challenge to developers of biomedical knowledgebases. Very few knowledgebases have resources to scale to the whole relevant literature and all have to prioritize their efforts. In this work, we take a first step to alleviating the lack of curator time in RNA science by generating summaries of literature for noncoding RNAs using large language models (LLMs). We demonstrate that high-quality, factually accurate summaries with accurate references can be automatically generated from the literature using a commercial LLM and a chain of prompts and checks. Manual assessment was carried out for a subset of summaries, with the majority being rated extremely high quality. We apply our tool to a selection of >4600 ncRNAs and make the generated summaries available via the RNAcentral resource. We conclude that automated literature summarization is feasible with the current generation of LLMs, provided that careful prompting and automated checking are applied. Database URL: https://rnacentral.org/.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
A.B. is an editor at DATABASE but was not involved in the editorial process of this manuscript.
Figures



References
-
- Joachimiak MP, Caufield JH, Harris NL et al. Gene set summarization using large language models. ArXiv 2023.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

