Microglial cGAS Deletion Preserves Intercellular Communication and Alleviates Amyloid-β-Induced Pathogenesis of Alzheimer's Disease
- PMID: 39908354
- PMCID: PMC11948024
- DOI: 10.1002/advs.202410910
Microglial cGAS Deletion Preserves Intercellular Communication and Alleviates Amyloid-β-Induced Pathogenesis of Alzheimer's Disease
Abstract
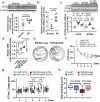
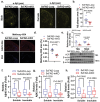
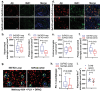
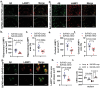
Innate immune activation plays a crucial role in the pathogenesis of Alzheimer's disease (AD) and related dementias (ADRD). The cytosolic DNA sensing pathway, involving cGAMP synthase (cGAS) and Stimulator of Interferon Genes (STING), has emerged as a key mediator of neurodegenerative diseases. However, the precise mechanisms through which cGAS activation influences AD progression remain poorly understood. In this study, we observed significant up-regulation of cGAS-STING signaling pathway in AD. Notably, this increase is primarily attributed to microglia, rather than non-microglial cell types. Using an inducible, microglia-specific cGAS knockout mouse model in the 5xFAD background, we demonstrated that deleting microglial cGAS at the onset of amyloid-β (Aβ) pathology profoundly restricts plaque accumulation and protects mice from Aβ-induced cognitive impairment. Mechanistically, our study revealed cGAS promotes plaque-associated microglia accumulation and is essential for inflammasome activation. Moreover, we showed that restricting cGAS-mediated innate immunity is crucial for preserving inter-cellular communication in the brain and induces pleiotrophin, a neuroprotective factor. These findings offer novel insights into the specific roles of the innate immune system in AD employing a cell-type-specific approach. The conclusions provide a foundation for targeted interventions to modulate the microglial cGAS-STING signaling pathway, offering promising therapeutic strategy for AD treatment.
Keywords: Alzheimer's disease; cGAS; innate immune; microglia.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Chen Q., Sun L., Chen Z. J., Nat. Immunol. 2016, 17, 1142. - PubMed
MeSH terms
Substances
Grants and funding
- T32 AG021890/AG/NIA NIH HHS/United States
- Methodist Hospital Foundation
- R01 AG085545/AG/NIA NIH HHS/United States
- P30 AG013319/AG/NIA NIH HHS/United States
- William and Ella Owens Medical Research Foundation
- Cure Alzheimer's Fund
- R01 AG061729/AG/NIA NIH HHS/United States
- T32AG021890/AG/NIA NIH HHS/United States
- RF1 AG061729/AG/NIA NIH HHS/United States
- University of Texas Health Science Center at San Antonio intramural institutional research funds
- P30 AG066546/AG/NIA NIH HHS/United States
- P30 AG044271/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
