RNA polymerase II coordinates histone deacetylation at active promoters
- PMID: 39908363
- PMCID: PMC11797538
- DOI: 10.1126/sciadv.adt3037
RNA polymerase II coordinates histone deacetylation at active promoters
Abstract
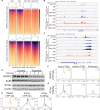
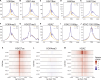
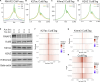
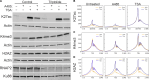
Nucleosomes at promoters of active genes are marked by specific histone post-translational modifications and histone variants. These features are thought to promote the formation and maintenance of an "open" chromatin environment that is suitable for transcription. However, recent reports have drawn conflicting conclusions about whether these histone modifications depend on active transcription. To further interrogate this relationship, we inhibited transcription initiation using triptolide, which triggered degradation of RNA polymerase II, and examined the impact on histone modifications. Transcription initiation was not required for either hormone-induced or steady-state active histone modifications at transcription start sites (TSSs) and enhancers. Rather, blocking transcription initiation increased the levels of histone acetylation and H2AZ incorporation at active TSSs. P300 activity was dispensable for this effect, but inhibition of histone deacetylases masked the increased acetylation. Together, our results demonstrate that active histone modifications occur independently of transcription. Furthermore, our findings suggest that the process of transcription coordinates the removal of these modifications to limit gene activity.
Figures
Update of
-
RNA Polymerase II coordinates histone deacetylation at active promoters.bioRxiv [Preprint]. 2024 Sep 17:2024.09.17.613553. doi: 10.1101/2024.09.17.613553. bioRxiv. 2024. Update in: Sci Adv. 2025 Feb 07;11(6):eadt3037. doi: 10.1126/sciadv.adt3037. PMID: 39345547 Free PMC article. Updated. Preprint.
References
-
- Talbert P. B., Henikoff S., The yin and yang of histone marks in transcription. Annu. Rev. Genomics Hum. Genet. 22, 147–170 (2021). - PubMed
-
- Wang Z., Chivu A. G., Choate L. A., Rice E. J., Miller D. C., Chu T., Chou S. P., Kingsley N. B., Petersen J. L., Finno C. J., Bellone R. R., Antczak D. F., Lis J. T., Danko C. G., Prediction of histone post-translational modification patterns based on nascent transcription data. Nat. Genet. 54, 295–305 (2022). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

