A multi-ancestry genome-wide association study and evaluation of polygenic scores of LDL-C levels
- PMID: 39909172
- PMCID: PMC11927683
- DOI: 10.1016/j.jlr.2025.100752
A multi-ancestry genome-wide association study and evaluation of polygenic scores of LDL-C levels
Abstract
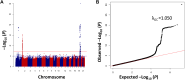
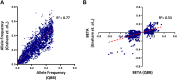
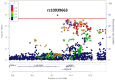
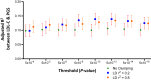
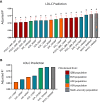
The genetic determinants of low-density lipoprotein cholesterol (LDL-C) levels in blood have been predominantly explored in European populations and remain poorly understood in Middle Eastern populations. We investigated the genetic architecture of LDL-C variation in Qatar by conducting a genome-wide association study (GWAS) on serum LDL-C levels using whole genome sequencing data of 13,701 individuals (discovery; n = 5,939, replication; n = 7,762) from the population-based Qatar Biobank (QBB) cohort. We replicated 168 previously reported loci from the largest LDL-C GWAS by the Global Lipids Genetics Consortium (GLGC), with high correlation in allele frequencies (R2 = 0.77) and moderate correlation in effect sizes (Beta; R2 = 0.53). We also performed a multi-ancestry meta-analysis with the GLGC study using MR-MEGA (Meta-Regression of Multi-Ethnic Genetic Association) and identified one novel LDL-C-associated locus; rs10939663 (SLC2A9; genomic control-corrected P = 1.25 × 10-8). Lastly, we developed Qatari-specific polygenic score (PGS) panels and tested their performance against PGS derived from other ancestries. The multi-ancestry-derived PGS (PGS000888) performed best at predicting LDL-C levels, whilst the Qatari-derived PGS showed comparable performance. Overall, we report a novel gene associated with LDL-C levels, which may be explored further to decipher its potential role in the etiopathogenesis of cardiovascular diseases. Our findings also highlight the importance of population-based genetics in developing PGS for clinical utilization.
Keywords: LDL; atherosclerosis; cholesterol; dyslipidemias; genomics.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
-
- Ference B.A., Ginsberg H.N., Graham I., Ray K.K., Packard C.J., Bruckert E., et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 2017;38:2459–2472. - PMC - PubMed
-
- Okrainec K., Banerjee D.K., Eisenberg M.J. Coronary artery disease in the developing world. Am. Heart J. 2004;148:7–15. - PubMed
-
- Baigent C., Keech A., Kearney P.M., Blackwell L., Buck G., Pollicino C., et al. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet. 2005;366:1267–1278. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

