Impaired primitive erythropoiesis and defective vascular development in Trim71-KO embryos
- PMID: 39909558
- PMCID: PMC11799773
- DOI: 10.26508/lsa.202402956
Impaired primitive erythropoiesis and defective vascular development in Trim71-KO embryos
Abstract
The transition of an embryo from gastrulation to organogenesis requires precisely coordinated changes in gene expression, but the underlying mechanisms remain unclear. The RNA-binding protein Trim71 is essential for development and serves as a potent regulator of post-transcriptional gene expression. Here, we show that global deficiency of Trim71 induces severe defects in mesoderm-derived cells at the onset of organogenesis. Murine Trim71-KO embryos displayed impaired primitive erythropoiesis, yolk sac vasculature, heart function, and circulation, explaining the embryonic lethality of these mice. Tie2 Cre Trim71 conditional knockout did not induce strong defects, showing that Trim71 expression in endothelial cells and their immediate progenitors is dispensable for embryonic survival. scRNA-seq of E7.5 global Trim71-KO embryos revealed that transcriptomic changes arise already at gastrulation, showing a strong up-regulation of the mesodermal pioneer transcription factor Eomes. We identify Eomes as a direct target of Trim71-mediated mRNA repression via the NHL domain, demonstrating a functional link between these important regulatory genes. Taken together, our data suggest that Trim71-dependent control of gene expression at gastrulation establishes a framework for proper development during organogenesis.
© 2025 Beckröge et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
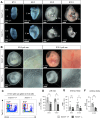



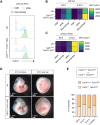

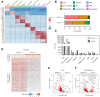



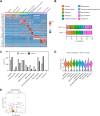


Similar articles
-
Heat shock protein 60 regulates yolk sac erythropoiesis in mice.Cell Death Dis. 2019 Oct 10;10(10):766. doi: 10.1038/s41419-019-2014-2. Cell Death Dis. 2019. PMID: 31601784 Free PMC article.
-
The RNA hairpin binder TRIM71 modulates alternative splicing by repressing MBNL1.Genes Dev. 2019 Sep 1;33(17-18):1221-1235. doi: 10.1101/gad.328492.119. Epub 2019 Aug 1. Genes Dev. 2019. PMID: 31371437 Free PMC article.
-
TRIM71 binds to IMP1 and is capable of positive and negative regulation of target RNAs.Cell Cycle. 2020 Sep;19(18):2314-2326. doi: 10.1080/15384101.2020.1804232. Epub 2020 Aug 20. Cell Cycle. 2020. PMID: 32816599 Free PMC article.
-
Eomesodermin-At Dawn of Cell Fate Decisions During Early Embryogenesis.Curr Top Dev Biol. 2017;122:93-115. doi: 10.1016/bs.ctdb.2016.09.001. Epub 2016 Oct 13. Curr Top Dev Biol. 2017. PMID: 28057273 Review.
-
Molecular and biological functions of TRIM-NHL RNA-binding proteins.Wiley Interdiscip Rev RNA. 2021 Mar;12(2):e1620. doi: 10.1002/wrna.1620. Epub 2020 Aug 1. Wiley Interdiscip Rev RNA. 2021. PMID: 32738036 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
