Kernel-bounded clustering for spatial transcriptomics enables scalable discovery of complex spatial domains
- PMID: 39909714
- PMCID: PMC11874963
- DOI: 10.1101/gr.278983.124
Kernel-bounded clustering for spatial transcriptomics enables scalable discovery of complex spatial domains
Abstract
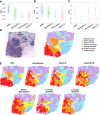
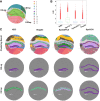
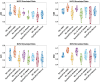
Spatial transcriptomics are a collection of technologies that have enabled characterization of gene expression profiles and spatial information in tissue samples. Existing methods for clustering spatial transcriptomics data have primarily focused on data transformation techniques to represent the data suitably for subsequent clustering analysis, often using an existing clustering algorithm. These methods have limitations in handling complex data characteristics with varying densities, sizes, and shapes (in the transformed space on which clustering is performed), and they have high computational complexity, resulting in unsatisfactory clustering outcomes and slow execution time even with GPUs. Rather than focusing on data transformation techniques, we propose a new clustering algorithm called kernel-bounded clustering (KBC). It has two unique features: (1) It is the first clustering algorithm that employs a distributional kernel to recruit members of a cluster, enabling clusters of varying densities, sizes, and shapes to be discovered, and (2) it is a linear-time clustering algorithm that significantly enhances the speed of clustering analysis, enabling researchers to effectively handle large-scale spatial transcriptomics data sets. We show that (1) KBC works well with a simple data transformation technique called the Weisfeiler-Lehman scheme, and (2) a combination of KBC and the Weisfeiler-Lehman scheme produces good clustering outcomes, and it is faster and easier-to-use than many methods that employ existing clustering algorithms and data transformation techniques.
© 2025 Zhang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures







Similar articles
-
DGSIST: Clustering spatial transcriptome data based on deep graph structure Infomax.Methods. 2024 Nov;231:226-236. doi: 10.1016/j.ymeth.2024.10.002. Epub 2024 Oct 15. Methods. 2024. PMID: 39413889
-
STGNNks: Identifying cell types in spatial transcriptomics data based on graph neural network, denoising auto-encoder, and k-sums clustering.Comput Biol Med. 2023 Nov;166:107440. doi: 10.1016/j.compbiomed.2023.107440. Epub 2023 Sep 9. Comput Biol Med. 2023. PMID: 37738898
-
BFAST: joint dimension reduction and spatial clustering with Bayesian factor analysis for zero-inflated spatial transcriptomics data.Brief Bioinform. 2024 Sep 23;25(6):bbae594. doi: 10.1093/bib/bbae594. Brief Bioinform. 2024. PMID: 39552067 Free PMC article.
-
A comprehensive survey of dimensionality reduction and clustering methods for single-cell and spatial transcriptomics data.Brief Funct Genomics. 2024 Dec 6;23(6):733-744. doi: 10.1093/bfgp/elae023. Brief Funct Genomics. 2024. PMID: 38860675 Review.
-
Computational Strategies and Algorithms for Inferring Cellular Composition of Spatial Transcriptomics Data.Genomics Proteomics Bioinformatics. 2024 Sep 13;22(3):qzae057. doi: 10.1093/gpbjnl/qzae057. Genomics Proteomics Bioinformatics. 2024. PMID: 39110523 Free PMC article. Review.
References
-
- Aggarwal CC. 2015. Data mining: the textbook, Vol. 1. Springer, Cham, Switzerland.
-
- Arthur D, Vassilvitskii S. 2006. How slow is the k-means method? In SCG '06: Proceedings of the twenty-second annual symposium on Computational Geometry, Sedona, AZ, pp. 144–153. 10.1145/1137856.1137880 - DOI
-
- Bandaragoda TR, Ting KM, Albrecht D, Liu FT, Zhu Y, Wells JR. 2018. Isolation-based anomaly detection using nearest-neighbor ensembles. Comput Intell 34: 968–998. 10.1111/coin.12156 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
