Brd7 loss reawakens dormant metastasis initiating cells in lung by forging an immunosuppressive niche
- PMID: 39910049
- PMCID: PMC11799300
- DOI: 10.1038/s41467-025-56347-2
Brd7 loss reawakens dormant metastasis initiating cells in lung by forging an immunosuppressive niche
Abstract
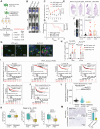
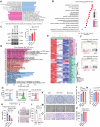
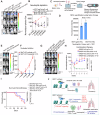
Metastasis in cancer is influenced by epigenetic factors. Using an in vivo screen, we demonstrate that several subunits of the polybromo-associated BAF (PBAF) chromatin remodeling complex, particularly Brd7, are required for maintaining breast cancer metastatic dormancy in the lungs of female mice. Brd7 loss induces metastatic reawakening, along with modifications in epigenomic landscapes and upregulated oncogenic signaling. Breast cancer cells harboring Brd7 inactivation also reprogram the surrounding immune microenvironment by downregulating MHC-1 expression and promoting a pro-metastatic cytokine profile. Flow cytometric and single-cell analyses reveal increased levels of pro-tumorigenic inflammatory and transitional neutrophils, CD8+ exhausted T cells, and CD4+ stress response T cells in lungs from female mice harboring Brd7-deficient metastases. Finally, attenuating this immunosuppressive milieu by neutrophil depletion, neutrophil extracellular trap (NET) inhibition, or immune checkpoint therapy abrogates metastatic outgrowth. These findings implicate Brd7 and PBAF in triggering metastatic outgrowth in cancer, pointing to targetable underlying mechanisms involving specific immune cell compartments.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: All the authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

