Sex-specific and cell-type-specific changes in chaperone-mediated autophagy across tissues during aging
- PMID: 39910244
- PMCID: PMC12003181
- DOI: 10.1038/s43587-024-00799-6
Sex-specific and cell-type-specific changes in chaperone-mediated autophagy across tissues during aging
Abstract
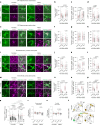
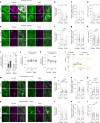
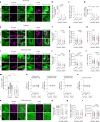
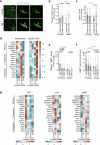
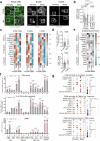
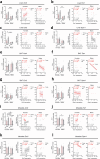
Aging leads to progressive decline in organ and tissue integrity and function, partly due to loss of proteostasis and autophagy malfunctioning. A decrease with age in chaperone-mediated autophagy (CMA), a selective type of lysosomal degradation, has been reported in various organs and cells from rodents and humans. Disruption of CMA recapitulates features of aging, whereas activating CMA in mice protects against age-related diseases such as Alzheimer's, retinal degeneration and/or atherosclerosis. However, sex-specific and cell-type-specific differences in CMA with aging remain unexplored. Here, using CMA reporter mice and single-cell transcriptomic data, we report that most organs and cell types show CMA decline with age, with males exhibiting a greater decline with aging. Reduced CMA is often associated with fewer lysosomes competent for CMA. Transcriptional downregulation of CMA genes may further contribute to CMA decline, especially in males. These findings suggest that CMA differences may influence organ vulnerability to age-related degeneration.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.M.C. is a co-founder and scientific advisor for the autophagy program at Life Biosciences. The other authors declare no competing interests relating to this work.
Figures











Similar articles
-
LAMP2A, and other chaperone-mediated autophagy related proteins, do not decline with age in genetically heterogeneous UM-HET3 mice.Aging (Albany NY). 2023 Jun 13;15(11):4685-4698. doi: 10.18632/aging.204796. Epub 2023 Jun 13. Aging (Albany NY). 2023. PMID: 37315291 Free PMC article.
-
Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome.Cell. 2021 May 13;184(10):2696-2714.e25. doi: 10.1016/j.cell.2021.03.048. Epub 2021 Apr 22. Cell. 2021. PMID: 33891876 Free PMC article.
-
Long-lived mice with reduced growth hormone signaling have a constitutive upregulation of hepatic chaperone-mediated autophagy.Autophagy. 2021 Mar;17(3):612-625. doi: 10.1080/15548627.2020.1725378. Epub 2020 Feb 12. Autophagy. 2021. PMID: 32013718 Free PMC article.
-
Chaperone-mediated autophagy: a gatekeeper of neuronal proteostasis.Autophagy. 2021 Aug;17(8):2040-2042. doi: 10.1080/15548627.2021.1935007. Epub 2021 Jun 10. Autophagy. 2021. PMID: 34110247 Free PMC article. Review.
-
Chaperone-mediated autophagy in neurodegenerative diseases: mechanisms and therapy.Mol Cell Biochem. 2023 Oct;478(10):2173-2190. doi: 10.1007/s11010-022-04640-9. Epub 2023 Jan 25. Mol Cell Biochem. 2023. PMID: 36695937 Review.
Cited by
-
Liver, ageing and disease.Nat Rev Gastroenterol Hepatol. 2025 Jul 28. doi: 10.1038/s41575-025-01099-z. Online ahead of print. Nat Rev Gastroenterol Hepatol. 2025. PMID: 40721658 Review.
References
MeSH terms
Grants and funding
- R01 AG021904/AG/NIA NIH HHS/United States
- AG054108/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- PID2021-126864NB I00/Ministerio de Ciencia Tecnología y Telecomunicaciones (Ministerio de Ciencia Tecnología y Telecomunicaciones de Costa Rica)
- R37 AG021904/AG/NIA NIH HHS/United States
- DK124308/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- RF1 AG054108/AG/NIA NIH HHS/United States
- DK098408/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- T32AG023475/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- R01 DK098408/DK/NIDDK NIH HHS/United States
- IRACDA-BETTR/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- T32 AG023475/AG/NIA NIH HHS/United States
- T32GM007491/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- N/A/JPB Foundation
- R01 DK124308/DK/NIDDK NIH HHS/United States
- F31 AG08419/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- T32 GM007491/GM/NIGMS NIH HHS/United States
- Margarita Salas Contract/Instituto Carlos Slim de la Salud (Carlos Slim Health Institute)
- T32GM007491/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- T32AG023475/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- AG031782/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- F31 AG084192/AG/NIA NIH HHS/United States
- AG021904/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- P01 AG031782/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

