Rapid and sensitive protein complex alignment with Foldseek-Multimer
- PMID: 39910251
- PMCID: PMC11903335
- DOI: 10.1038/s41592-025-02593-7
Rapid and sensitive protein complex alignment with Foldseek-Multimer
Abstract
Advances in computational structure prediction will vastly augment the hundreds of thousands of currently available protein complex structures. Translating these into discoveries requires aligning them, which is computationally prohibitive. Foldseek-Multimer computes complex alignments from compatible chain-to-chain alignments, identified by efficiently clustering their superposition vectors. Foldseek-Multimer is 3-4 orders of magnitudes faster than the gold standard, while producing comparable alignments; this allows it to compare billions of complex pairs in 11 h. Foldseek-Multimer is open-source software available at GitHub via https://github.com/steineggerlab/foldseek/ , https://search.foldseek.com/search/ and the BFMD database.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.S. acknowledges outside interest in Stylus Medicine. The remaining authors declare no competing interests.
Figures

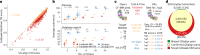
References
-
- Levy, E. D. & Teichmann, S. Structural, evolutionary, and assembly principles of protein oligomerization. Prog. Mol. Biol. Transl. Sci.117, 25–51 (2013). - PubMed
-
- Zhang, C., Shine, M., Pyle, A. M. & Zhang, Y. US-align: universal structure alignments of proteins, nucleic acids, and macromolecular complexes. Nat. Methods19, 1109–1115 (2022). - PubMed
MeSH terms
Substances
Grants and funding
- 2021-R1C1-C102065/National Research Foundation of Korea (NRF)
- 2021-M3A9-I4021220/National Research Foundation of Korea (NRF)
- RS-2024-00396026/National Research Foundation of Korea (NRF)
- RS-2023-00250470/National Research Foundation of Korea (NRF)
- Creative-Pioneering Researchers Program/Seoul National University
LinkOut - more resources
Full Text Sources

