STAT6 mutations compensate for CREBBP mutations and hyperactivate IL4/STAT6/RRAGD/mTOR signaling in follicular lymphoma
- PMID: 39910284
- PMCID: PMC11976298
- DOI: 10.1038/s41375-025-02525-6
STAT6 mutations compensate for CREBBP mutations and hyperactivate IL4/STAT6/RRAGD/mTOR signaling in follicular lymphoma
Abstract
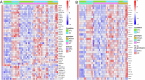
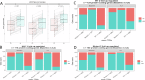
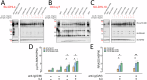
Activating mutations in STAT6 are common in Follicular Lymphoma (FL) and transformed FL and various other B cell lymphomas. Here, we report RNA-seq based gene expression data on normal human lymph node derived B lymphocytes (NBC; N = 6), and primary human FL WT (N = 11) or mutant (N = 4) for STAT6 before and after ex vivo stimulation with IL4. We found that STAT6 mutants result in broad based augmentation of IL4-induced gene expression. Unexpectedly, in FL with WT STAT6 we measured reduced baseline and IL4-induced gene expression levels when compared with NBC lymphocytes or FL with STAT6 mutations. We tracked the attenuated IL4/JAK/STAT6 response to co-existing CREBBP mutations and experimentally verified that intact CREBBP is required for the induction of many IL4-induced genes. One of the IL4-induced genes here identified is RRAGD, a small G-protein involved in lysosomal mTOR activation. We show that IL4 treatment induced RRAGD expression, that RRAGD is required for mTOR activation in lymphoma cells and that IL4-enhanced BCR signaling induced mTOR activation. The IL4 and BCR-induced mTOR activation was reduced by CREBBP mutants and augmented by mutant STAT6, establishing a link between STAT6 mutations and mTOR regulated pro-growth pathways in lymphoma.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Sami Malek owns stock in Abbvie. The other authors declared no conflicts.
Figures




References
-
- Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–69. - PubMed
-
- Cheung KJ, Johnson NA, Affleck JG, Severson T, Steidl C, Ben-Neriah S, et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010;70:9166–74. - PubMed
-
- Stevenson FK, Stevenson GT. Follicular lymphoma and the immune system: from pathogenesis to antibody therapy. Blood. 2012;119:3659–67. - PubMed
-
- Relander T, Johnson NA, Farinha P, Connors JM, Sehn LH, Gascoyne RD. Prognostic factors in follicular lymphoma. J Clin Oncol. 2010;28:2902–13. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

