Synthetic lethality of mRNA quality control complexes in cancer
- PMID: 39910291
- PMCID: PMC11864970
- DOI: 10.1038/s41586-024-08398-6
Synthetic lethality of mRNA quality control complexes in cancer
Abstract
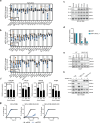
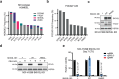
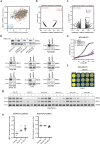
Synthetic lethality exploits the genetic vulnerabilities of cancer cells to enable a targeted, precision approach to treat cancer1. Over the past 15 years, synthetic lethal cancer target discovery approaches have led to clinical successes of PARP inhibitors2 and ushered several next-generation therapeutic targets such as WRN3, USP14, PKMYT15, POLQ6 and PRMT57 into the clinic. Here we identify, in human cancer, a novel synthetic lethal interaction between the PELO-HBS1L and SKI complexes of the mRNA quality control pathway. In distinct genetic contexts, including 9p21.3-deleted and high microsatellite instability (MSI-H) tumours, we found that phenotypically destabilized SKI complex leads to dependence on the PELO-HBS1L ribosomal rescue complex. PELO-HBS1L and SKI complex synthetic lethality alters the normal cell cycle and drives the unfolded protein response through the activation of IRE1, as well as robust tumour growth inhibition. Our results indicate that PELO and HBS1L represent novel therapeutic targets whose dependence converges upon SKI complex destabilization, a common phenotypic biomarker in diverse genetic contexts representing a significant population of patients with cancer.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: V.P., A.E.R., K.S., S.K., K.K., D.C., D.W., P.L., S.P., M.T., A.C., S.W., M.B.P., D.M., V.M., X.S., C.L., S.S., Z.D., G.B., D.V., B.B., J.A.M., W.N.P. and J.P.P. are employees of AbbVie.
Figures










References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

