Antibody prophylaxis may mask subclinical SIV infections in macaques
- PMID: 39910294
- PMCID: PMC11882457
- DOI: 10.1038/s41586-024-08500-y
Antibody prophylaxis may mask subclinical SIV infections in macaques
Abstract
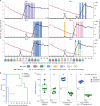
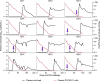
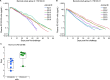
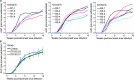
Broadly neutralizing antibodies (bNAbs) show potential to prevent human immunodeficiency virus (HIV-1) infection in humans1. However, there are limited data on the antibody concentrations required to prevent infection. Clinical trials of bNAb prophylaxis have demonstrated partial efficacy2, but the sampling frequency typically does not allow precise timing of infection events and concurrent antibody levels. Here, using simian immunodeficiency virus (SIV) infection of rhesus macaques, we show that although potent bNAbs can delay the onset of acute viremia, subclinical infections occur while bNAb levels remain high. Serial SIV challenge of monkeys given partially and fully neutralizing bNAbs revealed that 'viral blips'-low and transient plasma viremia-often occur while serum bNAb concentrations are well above currently accepted protective levels. To understand the precise timing of the infections resulting in such blips, we performed plasma viral sequencing on monkeys that were serially challenged with genetically barcoded SIV after bNAb administration. These analyses showed that subclinical infections occurred in most animals that were given potent bNAb prophylaxis. These subclinical infections occurred while antibody concentrations were 2- to 400-fold higher than the levels required to prevent fully viremic breakthrough infection. This study demonstrates that immunoprophylaxis can mask subclinical infections, which may affect the interpretation of prophylactic HIV-1 bNAb clinical trials.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Global HIV & AIDS Statistics — 2023 Fact Sheet. UNAIDSwww.unaids.org/en/resources/fact-sheet (2023).
-
- Landovitz, R. J., Scott, H. & Deeks, S. G. Prevention, treatment and cure of HIV infection. Nat. Rev. Microbiol.10.1038/s41579-023-00914-1 (2023). - PubMed
-
- Velloza, J., Delany-Moretlwe, S. & Baeten, J. M. Comprehensive HIV risk reduction interventions for 2020 and beyond: product choices and effective service-delivery platforms for individual needs and population-level impact. Curr. Opin. HIV AIDS14, 423–432 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

