The TB27 Transcriptomic Model for Predicting Mycobacterium tuberculosis Culture Conversion
- PMID: 39911144
- PMCID: PMC11792529
- DOI: 10.20411/pai.v10i1.770
The TB27 Transcriptomic Model for Predicting Mycobacterium tuberculosis Culture Conversion
Abstract
Rationale: Treatment monitoring of tuberculosis patients is complicated by a slow growth rate of Mycobacterium tuberculosis. Recently, host RNA signatures have been used to monitor the response to tuberculosis treatment.
Objective: Identifying and validating a whole blood-based RNA signature model to predict microbiological treatment responses in patients on tuberculosis therapy.
Methods: Using a multi-step machine learning algorithm to identify an RNA-based algorithm to predict the remaining time to culture conversion at flexible time points during anti-tuberculosis therapy.
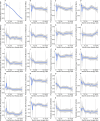
Results: The identification cohort included 149 patients split into a training and a test cohort, to develop a multistep algorithm consisting of 27 genes (TB27) for predicting the remaining time to culture conversion (TCC) at any given time. In the test dataset, predicted TCC and observed TCC achieved a correlation coefficient of r=0.98. An external validation cohort of 34 patients shows a correlation between predicted and observed days to TCC also of r=0.98.
Conclusion: We identified and validated a whole blood-based RNA signature (TB27) that demonstrates an excellent agreement between predicted and observed times to M. tuberculosis culture conversion during tuberculosis therapy. TB27 is a potential useful biomarker for anti-tuberculosis drug development and for prediction of treatment responses in clinical practice.
Keywords: biomarker; precision medicine; systems biology; therapy response; tuberculosis treatment.
Copyright © 2025 Pathogens and Immunity.
Conflict of interest statement
Stefan H.E. Kaufmann, Maja Reimann, Jan Heyckendorf, Sebastian Marwitz, and Christoph Lange declare that they are co-holders of patents on tuberculosis biomarkers
Figures



References
-
- World Health Organization. Global tuberculosis report 2023. Geneva, Switzerland: 2023.
-
- World Health Organization. Treatment of drug-susceptible tuberculosis: rapid communication. Geneva, Switzerland: WHO; 2021.
-
- World Health Organization. Rapid communication: key changes to the treatment of drug-resistant tuberculosis. Geneva, Switzerland: World Health Organization, 2022.
-
- Schaberg T, Brinkmann F, Feiterna-Sperling C, Geerdes-Fenge H, Hartmann P, Häcker B, Hauer B, Haas W, Heyckendorf J, Lange C.. Tuberkulose im Erwachsenenalter. Pneumologie. 2022;76(11):727–819. - PubMed
-
- Heyckendorf J, Georghiou SB, Frahm N, Heinrich N, Kontsevaya I, Reimann M, Holtzman D, Imperial M, Cirillo DM, Gillespie SH, Ruhwald M.. Tuberculosis Treatment Monitoring and Outcome Measures: New Interest and New Strategies. Clin Microbiol Rev. 2022;35(3):e0022721. doi: 10.1128/cmr.00227-21. PubMed PMID: 35311552; PMCID: PMC9491169. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
