High-purity CTC RNA sequencing identifies prostate cancer lineage phenotypes prognostic for clinical outcomes
- PMID: 39912912
- PMCID: PMC12046329
- DOI: 10.1158/2159-8290.CD-24-1509
High-purity CTC RNA sequencing identifies prostate cancer lineage phenotypes prognostic for clinical outcomes
Abstract
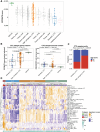
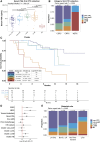
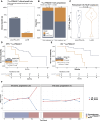
The development of treatment resistance remains universal for patients with metastatic prostate cancer, driven by AR alterations and lineage state transitions. Identifying the evolution of lineage transitions in treatment resistance has been limited by the challenges of collecting serial tissue biopsies on treatment, which can be overcome using blood-based liquid biopsies. Utilizing a novel circulating tumor cell (CTC) isolation approach, we collected 273 CTC samples from 117 patients with metastatic prostate cancer for RNA sequencing. 146 samples from 70 patients had tumor purity comparable to tissue biopsies. We identified four CTC transcriptional phenotypes, mirroring lineage states identified in tissue. Patients with a luminal-B-like CTC phenotype defined by persistent AR signaling and high proliferation, as well as those with a neuroendocrine CTC phenotype, had significantly shorter survival than patients with luminal-A-like and low proliferation phenotypes. In a prospective substudy, pre-treatment CTC luminal-B-like phenotype was associated with early progression on 177Lu-PSMA-617.
Conflict of interest statement
M.N. Sharifi reports grants from the Prostate Cancer Foundation (PCF), Department of Defense (DOD) and the National Institutes of Health (NIH) during the conduct of the study, and institutional research support from Novartis outside the submitted work. J.M. Sperger reports grants from the NIH during the conduct of the study. A.K. Taylor reports grants from PCF and DOD during the conduct of the study. J.L. Schehr reports grants from the NIH during the conduct of the study. K.T. Helzer reports employment of a family member with Epic Systems. M.L. Bootsma reports employment of his wife with Luminex, a biotechnology company that designs clinical assays. J.M. Floberg reports grants from the NIH during the conduct of the study. C.E. Kyriakopoulos reports grants and personal fees from Sanofi-Aventis, grants from AstraZeneca, Bristol Myers Squibb, Merck, and Madison Vaccines, and personal fees from AVEO Pharmaceuticals, Exelixis, Janssen, Pfizer, and Merck KGaA outside the submitted work. H. Emamekhoo reports personal fees from Bristol Myers Squibb, AVEO Pharmaceuticals, Janssen Biotech, Eisai, and Cardinal Health outside the submitted work. S.T. Tagawa reports grants from PCF during the conduct of the study; grants and personal fees from Johnson & Johnson, Pfizer, Gilead Sciences, Novartis, Telix Pharmaceuticals, Convergent Therapeutics, POINT, AstraZeneca, Lantheus, Bayer, Amgen, and Merck and personal fees from Regeneron outside the submitted work; and a patent for Biomarkers for Sacituzumab Govitecan therapy issued to Cornell/Gilead and a patent for Radiotherapeutic Conjugates for Treating Cancer pending to Cornell. M. Sjöström reports grants from the PCF and the Swedish Cancer Society during the conduct of the study, as well as personal fees from Astellas and Veracyte/Adelphi Targis outside the submitted work. A.D. Choudhury reports grants from Bayer, Eli Lilly and Company, and Sumitomo Pharma America, grants and personal fees from Pfizer, grants from Eli Lilly and Company, and personal fees from AstraZeneca, Astellas, Blue Earth Diagnostics, Janssen, Sanofi-Aventis, Tolmar, Lantheus, Daiichi Sankyo, and Boundless Bio outside the submitted work. A.J. Armstrong reports grants from the NIH during the conduct of the study, and grants and personal fees from Astellas, Pfizer, Janssen, Novartis AstraZeneca, and Bayer, grants from Bristol Myers Squibb, Merck, and Amgen, and personal fees from Myovant outside the submitted work. D.E. Rathkopf reports grants from the NIH during the conduct of the study, and uncompensated professional services and activities with AstraZeneca, Bayer, Bristol Myers Squibb, Genentech, Janssen Research & Development, LLC, Myovant Sciences, and Promontory Therapeutics Inc. H. Beltran reports other support from Pfizer, Amgen, Bayer, AstraZeneca, and Merck, grants and other support from Daiichi Sankyo and Novartis, and grants from Bristol Myers Squibb and Circle Pharma outside the submitted work. P.S. Nelson reports grants from PCF and the NIH during the conduct of the study, and grants from Janssen and personal fees from Bristol Myers Squibb, AstraZeneca, and Genentech outside the submitted work. F.Y. Feng reports nonfinancial support from Artera, Astellas, Bayer, Blue Earth Diagnostics, Bristol Meyers Squibb, ClearNote, Myovant, Roivant, Sanofi, Serimmune, and Amgen and personal fees and nonfinancial support from Janssen, Point Biopharma, and Novartis outside the submitted work. S.M. Dehm reports grants from the NCI and PCF during the conduct of the study; personal fees from Bristol Myers Squibb/Celgene and Oncternal Therapeutics outside the submitted work; and a patent for US-2013-0130241-A1 issued. X.X. Wei reports personal fees from Novartis outside the submitted work. R.R. McKay reports being a consultant and advisory for Ambrx, Arcus, AstraZeneca, AVEO Pharmaceuticals, Bayer, Blue Earth Diagnostics, Bristol Myers Squibb, Calithera, Caris, Dendreon, Daiichi Sankyo, Eli Lilly and Company, Eisai, Exelixis, Janssen, Merck, Myovant Sciences, Neomorph, Nimbus, Novartis, Pfizer, Sanofi, Seagen, Sorrento Therapeutics, Telix, and Tempus, as well as receiving institutional research support for AstraZeneca, Artera, Bayer, Bristol Myers Squibb, Exelixis, Oncternal Therapeutics, and Tempus. S.G. Zhao reports grants from the NIH and DOD during the conduct of the study; a patent for PORTOS and PAM50 Signatures in Prostate Cancer issued and licensed to Veracyte; employment of his spouse with Artera; and being a stockholder and a previous employee of Exact Sciences. J.M. Lang reports grants from the NIH, PCF, and DOD during the conduct of the study; personal fees from Janssen, Astellas, Arvinas, Gilead Sciences, Sanofi, Pfizer, AstraZeneca, Foundation Medicine, MacroGenics, Cytogen, and Cullgen and other support from Eolas outside the submitted work; and a patent for Circulating Tumor Cell Technology issued and licensed to Salus Discovery, LLC. No disclosures were reported by the other authors.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

