Campylobacter molothri sp. nov. isolated from wild birds
- PMID: 39913296
- PMCID: PMC11801493
- DOI: 10.1099/ijsem.0.006635
Campylobacter molothri sp. nov. isolated from wild birds
Abstract
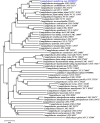
Twenty-nine hippuricase-positive Campylobacter strains were isolated from wild birds and river water. Previous characterization using atpA typing indicated that these strains were related to Campylobacter jejuni and Campylobacter coli but were most similar to three recently described hippuricase-positive Campylobacter species recovered from zebra finches, i.e. C. aviculae, C. estrildidarum and C. taeniopygiae. Phylogenetic analyses using 330 core genes placed the 29 strains into a clade well separated from the other Campylobacter taxa, indicating that these strains represent a novel Campylobacter species. Pairwise digital DNA-DNA hybridization and average nucleotide identity values were below 70 and 95 %, respectively, thus providing further supporting evidence of a novel taxon. Standard phenotypic testing was performed. All strains are microaerobic or anaerobic, motile, Gram-negative, spiral cells that are oxidase, catalase and nitrate reductase positive, but urease negative. Genomic analyses indicate that the 29 strains can potentially synthesize very few amino acids de novo and are auxotrophic for many amino acids and cofactors, similar to the species composing the Campylobacter lari group. In addition, these strains encode complete Entner-Doudoroff and Leloir pathways, suggesting that they may possess the ability to utilize both glucose and galactose; these pathways were also identified in the genomes of the zebra finch-associated taxa. The data presented here show that these strains represent a novel species within Campylobacter, for which the name Campylobacter molothri sp. nov. (type strain RM10537T=LMG 32306T=CCUG 75331T) is proposed.
Keywords: Campylobacter; blackbird; brown-headed cowbird; hippuricase; novel species.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


References
-
- Waldenstrom J, Griekspoor P. In: Campylobacter Ecology and Evolution. Meric G, Sheppard SK, editors. Norfolk, UK: Caister Academic Press; 2014. Ecology and host associations of Campylobacter in wild birds; pp. 265–286.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

