Comparisons of performances of structural variants detection algorithms in solitary or combination strategy
- PMID: 39913463
- PMCID: PMC11801633
- DOI: 10.1371/journal.pone.0314982
Comparisons of performances of structural variants detection algorithms in solitary or combination strategy
Abstract
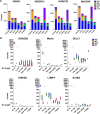
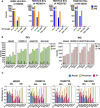
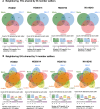
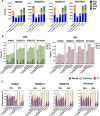
Structural variants (SVs) have been associated with changes in gene expression, which may contribute to alterations in phenotypes and disease development. However, the precise identification and characterization of SVs remain challenging. While long-read sequencing offers superior accuracy for SV detection, short-read sequencing remains essential due to practical and cost considerations, as well as the need to analyze existing short-read datasets. Numerous algorithms for short-read SV detection exist, but none are universally optimal, each having limitations for specific SV sizes and types. In this study, we evaluated the efficacy of six advanced SV detection algorithms, including the commercial software DRAGEN, using the GIAB v0.6 Tier 1 benchmark and HGSVC2 cell lines. We employed both individual and combination strategies, with systematic assessments of recall, precision, and F1 scores. Our results demonstrate that the union combination approach enhanced detection capabilities, surpassing single algorithms in identifying deletions and insertions, and delivered comparable recall and F1 scores to the commercial software DRAGEN. Interestingly, expanding the number of algorithms from three to five in the combination did not enhance performance, highlighting the efficiency of a well-chosen ensemble over a larger algorithmic pool.
Copyright: © 2025 Duan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
NO authors have competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

