A multi-modal fusion model with enhanced feature representation for chronic kidney disease progression prediction
- PMID: 39913621
- PMCID: PMC11801269
- DOI: 10.1093/bib/bbaf003
A multi-modal fusion model with enhanced feature representation for chronic kidney disease progression prediction
Abstract
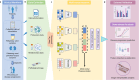
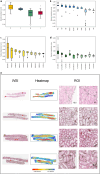
Artificial intelligence (AI)-based multi-modal fusion algorithms are pivotal in emulating clinical practice by integrating data from diverse sources. However, most of the existing multi-modal models focus on designing new modal fusion methods, ignoring critical role of feature representation. Enhancing feature representativeness can address the noise caused by modal heterogeneity at the source, enabling high performance even with small datasets and simple architectures. Here, we introduce DeepOmix-FLEX (Fusion with Learning Enhanced feature representation for X-modal or FLEX in short), a multi-modal fusion model that integrates clinical data, proteomic data, metabolomic data, and pathology images across different scales and modalities, with a focus on advanced feature learning and representation. FLEX contains a Feature Encoding Trainer structure that can train feature encoding, thus achieving fusion of inter-feature and inter-modal. FLEX achieves a mean AUC of 0.887 for prediction of chronic kidney disease progression on an internal dataset, exceeding the mean AUC of 0.727 using conventional clinical variables. Following external validation and interpretability analyses, our model demonstrated favorable generalizability and validity, as well as the ability to exploit markers. In summary, FLEX highlights the potential of AI algorithms to integrate multi-modal data and optimize the allocation of healthcare resources through accurate prediction.
Keywords: chronic kidney disease; computational pathology; deep learning; multi-modal; multi-omics; progression prediction.
© The Author(s) 2025. Published by Oxford University Press.
Figures



Similar articles
-
Transferable multi-modal fusion in knee angles and gait phases for their continuous prediction.J Neural Eng. 2023 May 24;20(3). doi: 10.1088/1741-2552/accd22. J Neural Eng. 2023. PMID: 37059084
-
MMGCN: Multi-modal multi-view graph convolutional networks for cancer prognosis prediction.Comput Methods Programs Biomed. 2024 Dec;257:108400. doi: 10.1016/j.cmpb.2024.108400. Epub 2024 Sep 6. Comput Methods Programs Biomed. 2024. PMID: 39270533
-
The importance of multi-modal imaging and clinical information for humans and AI-based algorithms to classify breast masses (INSPiRED 003): an international, multicenter analysis.Eur Radiol. 2022 Jun;32(6):4101-4115. doi: 10.1007/s00330-021-08519-z. Epub 2022 Feb 17. Eur Radiol. 2022. PMID: 35175381 Free PMC article.
-
Artificial intelligence in chronic kidney diseases: methodology and potential applications.Int Urol Nephrol. 2025 Jan;57(1):159-168. doi: 10.1007/s11255-024-04165-8. Epub 2024 Jul 25. Int Urol Nephrol. 2025. PMID: 39052168 Free PMC article. Review.
-
Artificial intelligence in traditional Chinese medicine: advances in multi-metabolite multi-target interaction modeling.Front Pharmacol. 2025 Apr 15;16:1541509. doi: 10.3389/fphar.2025.1541509. eCollection 2025. Front Pharmacol. 2025. PMID: 40303920 Free PMC article. Review.
Cited by
-
Future Designs of Clinical Trials in Nephrology: Integrating Methodological Innovation and Computational Power.Sensors (Basel). 2025 Aug 8;25(16):4909. doi: 10.3390/s25164909. Sensors (Basel). 2025. PMID: 40871773 Free PMC article. Review.
References
MeSH terms
Grants and funding
- KF2422-93/Open Project of National Key Laboratory of Oncology Systems Medicine
- GZNL2023A03001/Major Project of Guangzhou National Laboratory
- 2024Z229/Ningbo Science and Technology Innovation Yongjiang 2035 Project
- 2024020919/Ningbo major project for high-level medical and healthcare teams
- 32341019/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

