Using multiomic integration to improve blood biomarkers of major depressive disorder: a case-control study
- PMID: 39914267
- PMCID: PMC11848217
- DOI: 10.1016/j.ebiom.2025.105569
Using multiomic integration to improve blood biomarkers of major depressive disorder: a case-control study
Abstract
Background: Major depressive disorder (MDD) is a leading cause of disability, with a twofold increase in prevalence in women compared to men. Over the last few years, identifying molecular biomarkers of MDD has proven challenging, reflecting interactions among multiple environmental and genetic factors. Recently, epigenetic processes have been proposed as mediators of such interactions, with the potential for biomarker development.
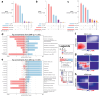
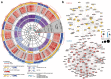
Methods: We characterised gene expression and two mechanisms of epigenomic regulation, DNA methylation (DNAm) and microRNAs (miRNAs), in blood samples from a cohort of individuals with MDD and healthy controls (n = 169). Case-control comparisons were conducted for each omic layer. We also defined gene coexpression networks, followed by step-by-step annotations across omic layers. Third, we implemented an advanced multiomic integration strategy, with covariate correction and feature selection embedded in a cross-validation procedure. Performance of MDD prediction was systematically compared across 6 methods for dimensionality reduction, and for every combination of 1, 2 or 3 types of molecular data. Feature stability was further assessed by bootstrapping.
Findings: Results showed that molecular and coexpression changes associated with MDD were highly sex-specific and that the performance of MDD prediction was greater when the female and male cohorts were analysed separately, rather than combined. Importantly, they also demonstrated that performance progressively increased with the number of molecular datasets considered.
Interpretation: Informational gain from multiomic integration had already been documented in other medical fields. Our results pave the way toward similar advances in molecular psychiatry, and have practical implications for developing clinically useful MDD biomarkers.
Funding: This work was supported by the Centre National de la Recherche Scientifique (contract UPR3212), the University of Strasbourg, the Université Sorbonne Paris Nord, the Université Paris Cité, the Fondation de France (FdF N° Engt:00081244 and 00148126; ECI, IY, RB, PEL), the French National Research Agency (ANR-18-CE37-0002, BE, CMC, ADD, PEL, ECI; ANR-18-CE17-0009, ADD; ANR-19-CE37-0010, PEL; ANR-21-RHUS-009, ADD, BE, CMC, CCB; ANR-22-PESN-0013, ADD), the Fondation pour la Recherche sur le Cerveau (FRC 2019, PEL), Fondation de France (2018, BE, CMC, ADD) and American Foundation for Suicide Prevention (AFSP YIG-1-102-19; PEL).
Keywords: DNA methylation; Depression; Multiomic integration; Sex differences; Transcriptomic; microRNA.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests BE received grants from ‘Agence Nationale de la Recherche (ANR)’ and consulting fees from Sanofi Winthrop. All other authors declare no competing interests.
Figures




References
-
- Marx W., Penninx B.W.J.H., Solmi M., et al. Major depressive disorder. Nat Rev Dis Primer. 2023;9:44. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

