Homologous recombination repair status in metastatic prostate cancer by next-generation sequencing and functional immunofluorescence
- PMID: 39914385
- PMCID: PMC11866514
- DOI: 10.1016/j.xcrm.2025.101937
Homologous recombination repair status in metastatic prostate cancer by next-generation sequencing and functional immunofluorescence
Abstract
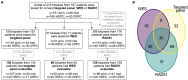
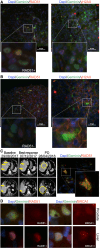
Metastatic prostate cancer (mPC) is enriched for homologous recombination repair (HRR) gene alterations, which have prognostic and predictive value. Routine clinical implementation of next-generation sequencing (NGS) is still limited. We investigated the association between genomic and functional loss of HRR, using NGS and RAD51 immunofluorescence (RAD51-IF) in 219 primary or metastatic biopsies from 187 patients with stage IV prostate cancer. NGS showed frequent genomic alterations in TP53 (40%), AR (15%), PTEN (14%), FOXA1 (12%), MYC (10%), BRCA2 (9%), ATM (8%), and BRCA1 (2%). We pursued RAD51-IF in 206 samples; of those, 139/206 (67%) were evaluable. 21% of samples had RAD51-low score compatible with HRR deficiency (HRD). RAD51-IF showed high sensitivity (71%) and specificity (86%) for BRCA1/2 alterations. Patients with RAD51-low scores experienced longer progression-free survival (PFS) on poly(ADP-ribose) polymerase inhibitors (PARPi) or platinum chemotherapy. RAD51-IF is feasible in routine clinical samples from patients with mPC and is associated with clinically relevant HRR gene alterations.
Keywords: BRCA2; HRR; PARPi; genomics; immunofluorescence; precision medicine; prostate cancer.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.M.-B. reports serving in an advisory role for MSD, Pfizer, Merck, Janssen, and Astellas Pharma and receiving honoraria or travel expenses from Roche, Sanofi Aventis, Astellas, Janssen, MSD, Bayer, Merck, and Pfizer. A.L.-G. and V.S. are co-inventors of a patent related to this work (WO2019122411A1). A.L.-G. is a current employee of AstraZeneca. V.S. has served as an advisor for GSK. J.M. has served as an advisor for AstraZeneca, Amunix/Sanofi, Daichii Sankyo, Janssen, MSD, Pfizer, and Roche; he is a member of the scientific board for Nuage Therapeutics and is involved as an investigator in several pharma-sponsored clinical trials, none of them related to this work.
Figures



References
-
- Abida W., Armenia J., Gopalan A., Brennan R., Walsh M., Barron D., Danila D., Rathkopf D., Morris M., Slovin S., et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis. Oncol. 2017;2017:1–16. doi: 10.1200/PO.17.00029. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

