Endothelial SHANK3 regulates tight junctions in the neonatal mouse blood-brain barrier through β-Catenin signaling
- PMID: 39915488
- PMCID: PMC11802743
- DOI: 10.1038/s41467-025-56720-1
Endothelial SHANK3 regulates tight junctions in the neonatal mouse blood-brain barrier through β-Catenin signaling
Abstract
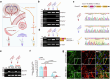
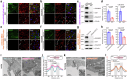
Autism spectrum disorder (ASD) is a neurodevelopmental disability condition arising from a combination of genetic and environmental factors. Despite the blood-brain barrier (BBB) serving as a crucial gatekeeper, conveying environmental influences into the brain parenchyma, the contributions of BBB in ASD pathogenesis remain largely uncharted. Here we report that SHANK3, an ASD-risk gene, expresses in the BBB-forming brain endothelial cells (BECs) and regulates tight junctional (TJ) integrity essential for BBB's barrier function. Endothelium-specific Shank3 (eShank3) knockout (KO) neonatal mice exhibit male-specific BBB-hyperpermeability, reduced neuronal excitability, and impaired ultra-sonic communications. Although BBB permeability is restored during adult age, the male mutant mice display reduced neuronal excitability and impaired sociability. Further analysis reveals that the BBB-hyperpermeability is attributed to the β-Catenin imbalance triggered by eShank3-KO. These findings highlight a pathogenic mechanism stemming from the ASD-risk Shank3, emphasizing the significance of neonatal BECs in the BBB as a potential therapeutic target for ASD.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Huber, J. D., Egleton, R. D. & Davis, T. P. Molecular physiology and pathophysiology of tight junctions in the blood-brain barrier. Trends Neurosci.24, 719–725 (2001). - PubMed
-
- Ouellette, J. et al. Vascular contributions to 16p11.2 deletion autism syndrome modeled in mice. Nat. Neurosci.23, 1090–1101 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

